Significance of Vibrio species in the marine organic carbon cycle— A review
Xiaohua ZHANG1'2*, Heyu LIN1, Xiaolei WANG1 & Brian AUSTIN3
1 College of Marine Life Sciences, Ocean University of China, Qingdao 266003, China; 2 Laboratory for Marine Ecology and Environmental Science, Qingdao National Laboratory for Marine Science and Technology,Qingdao 266071, China; 3 Institute of Aquaculture, University of Stirling, Stirling FK9 4LA, UK Received November 3, 2017; revised April 17, 2018; accepted May 29, 2018; published online August 6, 2018
Abstract
The genus Vibrio, belonging to Gammaproteobacteria of the phylum Proteobacteria, is a genetically and ecologically diverse group of heterotrophic bacteria, that are ubiquitous in marine environments, especially in coastal areas. In particular, vibrios dominate, i.e. up to 10% of the readily culturable marine bacteria in these habitats. The distribution of Vibrio spp. is shaped by various environmental parameters, notably temperature, salinity and dissolved organic carbon. Vibriospp. may utilize a wide range of organic carbon compounds, including chitin (this may be metabolized by most Vibrio spp.), alginic acid and agar. Many Vibrio spp. have very short replication times (as short as ~10 min), which could facilitate them developing into high biomass content albeit for relatively short durations. Although Vibriospp. usually comprise a minor portion (typically ~l%of the total bacterioplankton in coastal waters) of the total microbial population, they have been shown to proliferate explosively in response to various nutrient pulses, e.g., organic nutrients from algae blooms and iron from Saharan dust. Thus, Vibrio spp. may exert large impacts on marine organic carbon cycling especially in marginal seas. Genomics and related areas of investigation will reveal more about the molecular components and mechanisms involved in Vibrio-mediated biotransformation and remineralization processes.
Keywords Vibrio, Ecology, Carbon cycle, Marine, Organic carbon
1. Introduction
Organic matter remineralization by heterotrophic microorganisms is a central component of the marine carbon cycle. These microorganisms process approximately half of all CO2 initially fixed into organic carbon by phytoplankton, transforming, repackaging, and respiring organic carbon and simultaneously regenerating nutrients (Azam and Malfatti, 2007; Arnosti, 2014). Therefore, the variations of abundance, community structure and activities of heterotrophic microbial communities have important impacts on the mar-Corresponding author (email: xhzhang@ouc.edu.cn) ine carbon budget. Despite the importance of microbially driven carbon cycling, the specific factors that determine the extent, rate, and location of organic matter remineralization in the ocean are poorly understood.
The genus Vibrio is one of the best model marine heterotrophic bacterial groups. The organisms are widely distributed in the marine environment and in comparison, to other marine heterotrophic bacteria, many Vibrio species grow very fast with much shorter generation-times. For example, Vibrio parahaemolyticus has a shorter generation time of only ~10 min (Fujino et al., 1951; Sakazaki et al., 1963; Joseph et al., 1982) compared to Alteromonas with ~1 h generation time (Feller et al., 1994), Roseobacter with earth.scichina.com link.springer.com~1.3 h (Bruhn et al., 2005), and SAR11 which needs ~40 h to multiply (Simu and Hagstrom, 2004). The major phenotypic properties of Vibrio spp. include the presence of straight or curved chemo-organotrophic Gram-negative rods, that are motile by means of polar flagella, and require Na+ for growth (Farmer et al., 2005). Some Vibrio spp, e.g. V campbellii, are bioluminescent, a process which is controlled by quorum sensing (Gomez-Gil et al., 2014).
Many Vibrio spp. are well-known bacterial pathogens, causing disease in humans or marine animals. Vibrio cho- lerae, which was first described by Pacini in 1854, is the causative agent of human epidemic cholera (Farmer et al., 2005); V. parahaemolyticus causes severe gastroenteritis in humans through consumption of undercooked or contaminated seafood; V vulnificus cause severe bacteremia, skin and soft tissue infection through exposure of wounds to warm seawater (Farmer et al., 2005; Phillips and Satchell, 2017). Vibrio anguillarum, V harveyi, V campbellii, V al- ginolyticus and V. parahaemolyticus cause serious disease and thus severe economic losses to the aquaculture industry, worldwide (Farmer et al., 2005; Austin and Zhang, 2006; Hickey and Lee, 2017). In addition, V coralliilyticus may cause severe disease in corals, although other vibrios are part of the normal microbiota of coral species (Chimetto Tonon et al., 2017; Rubio-Portillo et al., 2018).
Over the past 40 years, many nonpathogenic species of Vibrio have also been described, including V diazotrophicus (a nitrogen-fixing species; Guerinot et al., 1982), V[=Alii- vibrio] fischeri (the bioluminescent symbiont of the squid Euprymna scolopes; Beijerinck, 1889; Lehmann and Neumann, 1896; Ruby et al., 2005), V. atypicus (associated with healthy shrimp; Wang et al., 2010), and V marisflavi (isolated from seawater; Wang et al., 2011).
Vibrios are ubiquitous in estuarine and marine environment on a global scale, especially in coastal areas (Thompson et al., 2006). They are easily cultured on standard (e.g. marine 2216E agar) and selective media (e.g. thiosulfate citrate bile salt sucrose agar) and are capable of a diverse array of metabolic activities (Thompson et al., 2006). Also, vibrios are capable of responding rapidly to nutrient pulses with explosive growth responses in amended microcosms (Martinez et al., 2013), such as during phytoplankton blooms (Gilbert et al., 2012) and dust storms (Westrich et al., 2016), suggesting that the short period of Vibrio blooms should be considered when attempting to determine their overall contribution to the recycling of organic macromolecules. Currently, the role of Vibrio spp. in marine organic carbon cycling, particularly in coastal environments, is likely to be underestimated (Thompson and Polz, 2006; Takemura et al., 2014).
The purpose of this review is to provide an overview of distribution and environmental drivers of Vibrio populations in the marine environment and to discuss their potential role in marine organic carbon cycling.
2.Dynamics of Vibrio populations in the marine environment
2.1Abundance
Vibrio population are widely distributed in estuarine and marine environments, and exhibit either of two alternative growth strategies: (1) free-living bacterioplankton; or (2) association with marine particles and/or living hosts, where they may act as mutual symbionts or disease agents (Thompson and Polz, 2006).
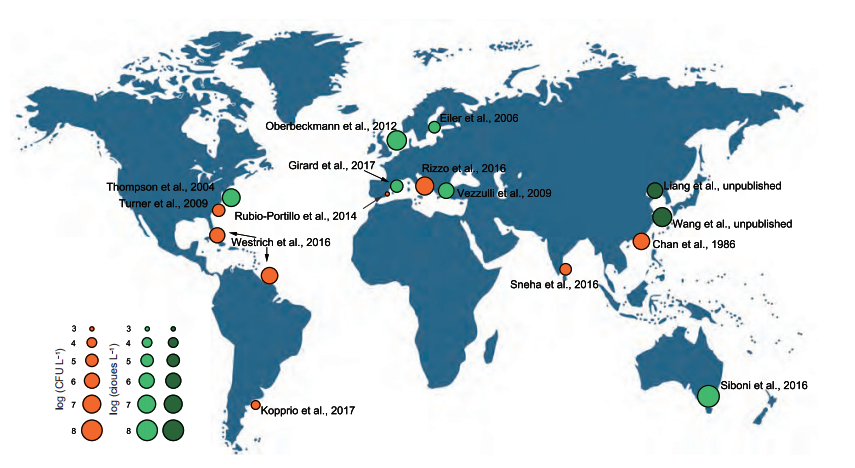
Vibrio spp. have been easily cultured on standard non- selective and selective media and thus were highly visible in the pre-molecular era of microbial ecology. Typically, Vibrio spp. comprise up to 10% of the readily culturable marine bacteria in culture-based studies (Eilers et al., 2000), with an average abundance of 103 to 106 CFU L-1 in estuarine and coastal waters (Figure 1). In contrast, with culture-independent methods, Vibrio population are generally ~1% of the total bacterioplankton in coastal waters (Thompson and Polz, 2006), with an average abundance of 104 to 108 16S rRNA copies L-1 in estuarine and coastal waters (Figure 1). Also, Vibrio spp. have been shown repeatedly to be present in high densities in and on marine organisms, such as fish, shrimp, mollusks, corals, sponges, zooplankton, algae, and seagrass (Thompson and Polz, 2006).
2.2Diversity of Vibrio populations
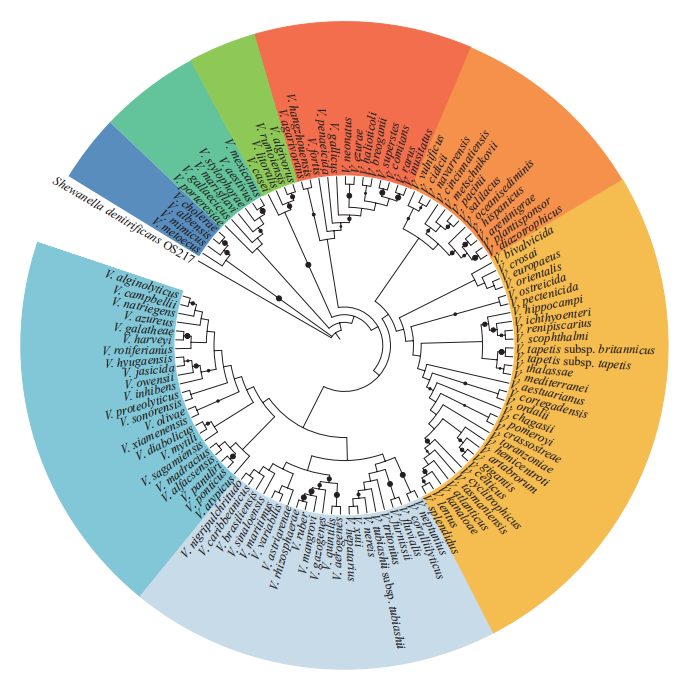
More than 110 Vibrio species have been described to date (Figure 2), and there are likely to be many more recognized in the future. However, different Vibrio spp. may exist in separate marine environments. To date, the diversity of Vibrio populations in marine environments has been examined largely on the basis of studying isolated Vibrio strains (Tall et al., 2013; Amin et al., 2016). Yet, the approach of using 16S rRNA gene similarity as the main phylogenetic species marker has failed with the genus Vibrio, because of the low interspecies resolution. Multilocus sequence analysis (MLSA) and other new phylogenetic markers (e.g., ferric uptake regulator gene) have been used as alternative methods (Thompson et al., 2005; Pascual et al., 2010; Machado and Gram, 2015). Recently, the diversity of Vibrio populations was investigated using environmental samples instead of isolated strains (e.g., Siboni et al., 2016). Here, DNA was extracted from seawater and sequenced by high-throughput methods using Vibrio-specific 16S rRNA gene primers that can intuitively and efficiently record Vibrio community diversity.
2.3Impact of environmental factors

Figure 1 Abundance of Vibrio spp. in different marine environments. Orange colour circles, by culture-based methods; green and dark green (unpublished data from our laboratory) colour circles, by culture-independent methods.
The abundance and community composition of planktonic Vibrio populations in marine environments are shaped by depth, dissolved oxygen, and turbidity), inorganic and organic nutrients (e.g., ammonium, nitrate, phosphate, silicate, and dissolved and particulate organic carbon), the carbon/ nitrogen ratio of suspended organic particulates, pigments (e.g., chlorophyll a and phaeopigment), predation by protozoans and viruses, and the abundance ofhost organisms, e.g., marine animals and algae (Takemura et al., 2014; Siboni et al., 2016; Davis et al., 2017; Kopprio et al., 2017; Kauffman et al., 2018). The numerous biotic and abiotic factors, and their interactions, increase the complexity of studies on Vibrio ecology.
The strongest environmental parameters that correlate with total Vibrio numbers are temperature and salinity. The abundance of vibrios increases along with heightened temperatures (2.5 to 32.5 C) and salinity (0 to 27 ppt, 1 ppt =10-12) (Takemura et al., 2014). It has been reported that the increase in dominance of Vibrio spp., within the plankton associated bacterial community of the North Sea during the last half century was correlated with increased sea surface temperature (Vezzulli et al., 2012). In fact, due to the strong relationship between the abundance of Vibrio and ambient environmental temperatures, it has been suggested that vibrios could serve as the microbial barometer of global warming or climate change (Vezzulli et al., 2012, 2016; Baker-Austin et al., 2017). Our study in the Changjiang Estuary showed that the abundance of Vibrio spp. increased from 10乂105 to 6乂105 cell mL-1 with the increase of salinity from 5.0 to 32.5 (data not shown). Most often, these two parameters explain the greatest variance in total (free-living and particle-associated) Vibrio abundance in the water column, although the magnitude of the correlation may depend on the parameter range examined (Takemura et al., 2014). Compared to salinity and temperature, other environmental parameters, including dissolved oxygen (DO), nitrogen, phosphate, pH, chlorophyll a and turbidity, usually explain less and inconsistent variance in total Vibrio numbers (Ta- kemura et al., 2014). However, some of the difficulty in making general statements regarding the relationship of Vibrio spp. to individual environmental parameters comes from the fact that their correlation may depend on the ranges examined (e.g., as for temperature and salinity), or inequality of the parameters that have been detected (Takemura et al., 2014; Davis et al., 2017).
In addition, dissolved organic carbon (DOC) has been reported to have a strong effect on Vibrio ecology (Takemura et al., 2014). DOC provides much of the nutrition needed by Vibrio spp. to live in estuarine and marine habitats. Vibrio spp. may incorporate, metabolize and produce organic matter, changing the chemical properties and bioavailability. Nevertheless, the bulk DOC that has been detected encompasses carbon derived from different sources and contains highly variable chemical compositions, which may be bioavailable to or preferable to different Vibrio spp. In fact, the chemical composition of DOC has rarely been analyzed in Vibrio ecology studies.

Figure 2 Phylogenetic tree of 115 Vibrio species. Neighbor-Joining phylogeny were reconstructed based on 16S rRNA gene sequences of the type strain of each species. The tree was rooted using Shewanella denitrificans OS217T as an outgroup. The branches were estimated from 1000 bootstrap replicates, and circles at internal nodes indicate bootstrap support greater than 50%; larger circles indicate greater bootstrap values.
Viruses are significant mortality agents of Vibrio spp. and have strong effects on Vibrio ecology. Recently, a major lineage of non-tailed dsDNA viruses infecting Vibrio spp. were recognized, and these viruses have a broad host range,killing on average 34 strains within four Vibrio spp. (Kauffman et al., 2018). Mortality of Vibrio spp. from viral lysis implies a substantial flux of DOC from cells into seawater, and this DOC may be taken up by other plankton thereby affecting pathways and rates of nutrient cycling. Therefore, the interaction between Vibrio and viruses may well be an important process to involve in carbon cycling.
Trends that apply to the whole Vibrio genus do not necessarily reflect those of individual species, in part due to species-specific associations with environmental factors and the hosts of Vibrio spp., including phyto- and zooplankton (Takemura et al., 2014). Total Vibrio populations and the well-studied pathogens, V cholerae, V parahaemolyticus, and V. vulnificus, correlate with shared and distinct environmental parameters. For V parahaemolyticus and V. vulnificus, temperature often explains more variance than does salinity in the same analysis, and for V. cholerae, diverse biotic variables, including specific phytoplankton and zooplankton taxa, may correlate more than abiotic parameters (Takemura et al., 2014; Davis et al., 2017; Kopprio et al., 2017). Unfortunately, biotic parameters, particularly individual plankton or zooplankton taxa, have been rarely studied, making these observations difficult to generalize (Takemura et al., 2014). Recently, it has been demonstrated that there is a significant correlation between the abundance of particle-associated Vibrio and phytoplankton community composition, with higher correlations to diatoms and raphi- dophytes than to dinoflagellates in the marine environment. This may be correlated with the bioavailable dissolved organic matter (DOM) released from the phytoplankton (Main et al., 2015).
Finally, Vibrio may enter into a dormant state, i.e., the viable but non-culturable state (VBNC; Xu et al., 1982), under unfavorable environmental conditions, e.g., low nutrient concentration, suboptimal or reduced temperature, elevated salinity, extreme pH and solar radiation. This physiological condition is a reversible state that ends with resuscitation when conditions once again become favorable (Oliver, 2010). However, the conditions leading to resuscitation of “dormant" cells are not always clear.
2.4 Vibrio blooms
Vibrio spp. are comparatively low abundant constituents in microbial assemblages in the water column (Thompson and Polz, 2006). However, in recent years, increasing evidence has shown that Vibrio populations are capable of blooming in the water column during which they may become the dominant members of the total bacterial population. This is in response to often poorly characterized abiotic or biotic factors. Several recent studies have shown that the increases in Vibrio abundance were associated with algal (such as diatom and phaeocystis) blooms (Miller et al., 2005; Gilbert et al., 2012) or with micronutrients, such as iron (Westrich et al., 2016). These blooms could have been missed previously because of their relatively short duration in the environment (Takemura et al., 2014).
The first reported Vibrio bloom was the so called “milky sea". Bioluminescent milky seas are characterized by continuous and substantial light emission from shelf or coastal surface waters that may span areas of thousands of square kilometers, and persist for days (Lapota et al., 1988; Miller et al., 2005). One case occurred in the northwestern Indian Ocean and was large enough (>17700 km2) to be detectable by satellite. This bloom was observed to glow over three consecutive nights (Miller et al., 2005). Evidence has implicated that the bioluminescent organism V harveyi (or its sister species such as V campbellii), in association with microalgae, namely the unicellular alga Phaeocystis globose, blooms in surface waters, and exerts a major role in the formation of milky seas (Lapota et al., 1988). However, the presumptive identification of V harveyi was based solely on phenotypic but not more modern and accurate molecular properties, which could have resulted in misidentification (Lapota et al., 1988).
Gilbert et al. (2012) observed an explosive Vibrio bloom at a temperate marine coastal site off Plymouth, UK, demonstrating the potential for rapid growth while part of a full marine bacterial community. In one month, a single unnamed Vibrio species, otherwise comprising only 0-2% of total 16S rRNA genes, multiplied to comprise 54% of the bacterial community一the largest bloom of any bacterial group observed over the course of a 6-year time series (Gilbert et al., 2012). Coincidently, there was a correlated bloom of the diatom Chaetoceros compressus, which is normally quite rare within the phytoplankton community. It is speculated that organic nutrients secreted by the unusually proliferating diatom taxon may have promoted the Vibrio bloom (Gilbert et al., 2012).
In addition, Westrich et al. (2016) demonstrated that Saharan dust nutrients promoted Vibrio bloom formation in marine surface waters of the Caribbean Sea and subtropical Atlantic Ocean. The arrival of Saharan dust in the Caribbean Sea and subtropical Atlantic Ocean coincided with high levels of dissolved iron that was followed by up to a 30-fold increase of culturable Vibrio over background levels within a 24 h period. Moreover, the relative abundance of Vibrio increased from ~1% to ~20% ofthe total microbial community (Westrich et al., 2016). Whether such blooms are rare or not remains unknown because sampling tends to be infrequent. However, observations made to date provide evidence that Vibrio are capable of rapid growth in the marine environment, especially in surface waters.
3.Vibrio-mediated degradation of organic matter
3.1 Organic carbon substrate
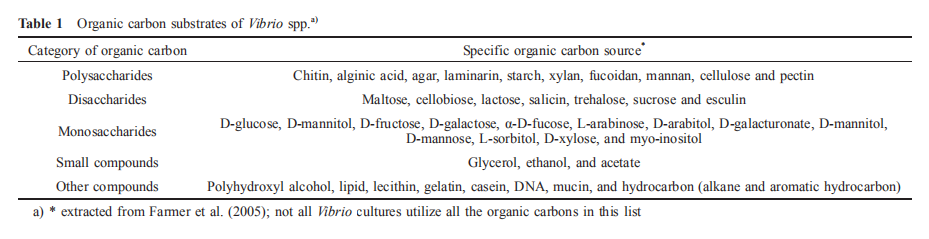
All currently described Vibrio spp. are obligate heterotrophs and, as such, rely on organic matter for their carbon sources. Generally, vibrios consume a wide range array of organic carbon compounds as carbon and energy sources, with most species being able to utilize over 40 compounds (Farmer et al., 2005). The list of organic carbon sources utilized include polysaccharides (e.g., chitin, alginic acid and agar), disaccharides (e.g., maltose, cellobiose and lactose), monosaccharides (e.g., D-glucose, D-mannitol and D-fructose), and other compounds, such as polyhydroxyl alcohols, alkanes, lipids and lecithin (Table 1). These lists were derived mainly from phenotypic tests carried out during conventional identification of novel Vibrio often by using commercial kits, such as API ZYM and Biolog MicroPlates.
Polysaccharides constitute a considerable fraction of dissolved and particulate organic matter in the oceans and are a central component of marine organic carbon cycles. Many of the polysaccharides are derived from macroalgal cell walls (i.e., alginic acid, agar, fucoidan and laminarin) or zooplankton exoskeletons (i.e., chitin).

Chitin, which consists of p-1,4-linked N-acetyl glucosamine monomers, is one of the most abundant organic compounds in the marine environment. It has been estimated that 1011 tonnes of chitin (primarily in the form of zooplankton and crustacean exoskeletons, and diatom shells) are produced annually in marine ecosystems; this polymer must be continually remineralized by microorganisms to support sustained primary production in the oceans (Li and Roseman, 2004). Chitin has been estimated to account for as much as 10% of all marine bacterial production (Kirchman and White, 1999). Vibrio spp. are considered to be the most prevalent bacteria that degrade chitin, although chitin may also be attacked by many other marine bacteria. In our previous work, 56.8% of Vibrio cultures (330 out of 581 isolates) isolated from Chinese marginal seas possessed chitin degrading activities, whereas only 2.5% of other marine bacteria could degrade chitin (data not shown). Thus, Vibrio spp. capable of hydrolyzing polymers, such as chitin, may create important trophic links within bacterioplankton communities (Thompson and Polz, 2006).
Alginate, which is a major cell wall matrix polysaccharide of brown macroalgae (accounts for >50% of the dry weight in brown macroalgae), is also an abundant source of organic matter in the marine environment. Alginate consists of uronic acids glucuronate and mannuronate, which are (1,4)- linked and arranged in homo- or heteropolymeric blocks. In our screening programme, 11.2% of Vibrio cultures (65 out of 581 isolates) could degrade alginic acid (data not shown). Agar is a major component of the cell walls in red algae, and is composed of two polysaccharides, i.e. agarose and agar- opectin. Agarose is the major component, and is made up of a-(1,3) and p-(1,4)-linked agarobiose units, each consisting of a galactose and galactopyranose monomer. It has been reported that many vibrios have the ability to hydrolyze and metabolize agarose as a carbon and energy source (Zhang and Sun, 2007; Chi et al., 2012).
In addition, Vibrio spp. may metabolize oil-derived compounds, such as alkane, aromatic hydrocarbon and benzoate (West et al., 1984). Vibrio populations were found to become the dominant fraction of oil-associated microbial communities from the Deepwater Horizon oil spill in the Gulf of Mexico, both from sea-surface samples (>31% of the total bacterial population; Hamdan and Fulmer, 2011) and saltmarsh plants contaminated with oil mousse (57% of the total bacterial population; Liu and Liu, 2013).
3.2 Responses of Vibrio populations to nutrient pulses in amended microcosms
The affinity of Vibrio to organic carbon substrates is quite high, so that Vibrio spp. may grow under high-nutrient conditions, such as in animal guts, and phytoplankton or zooplankton microenvironments. Indeed, the half-saturation constant for glucose is 29 and 500 卩mol L 1 for V. natriegens and V. parahaemolyticus, respectively, compared to typical bulk seawater glucose concentrations ranging from <14 nmol L-1 to 2 卩mol L-1 (Thompson and Polz, 2006).
Vibrio population are capable of responding rapidly to nutrient pulses and explosive growth in amended microcosms (Martinez et al., 2013). In the microcosm supplemented with 100 卩mol L-1 glucose and 16 卩mol L-1 nitrate, the decrease in the relative abundance of Prochlorales and Ricketsiales was accompanied by a large increase in the abundance of Vibrionales (59% of the total bacterial population, compared to less than 1% in the control) in the Gammaproteobacteria class. Also, a dramatic increase in the abundance of Vibrionales (44%) was observed in the microcosm supplemented with 100 卩mol L-1 glucose, 16 卩mol L-1 nitrate and 1 卩mol L-1 methylphosphonate (MPn) compared to the controls (Martinez et al., 2013).
4.Organic carbon storage and products
4.1 Cellular carbohydrate composition
The cell wall of Vibrio spp. contains the same three lipopolysaccharide (LPS) elements found in other Gram-negative bacteria namely lipid A, core polysaccharide, and an O- polysaccharide side chain (Farmer et al., 2005). The lipid A portion consists of a p(1'-6)-linked glucosamine disaccharide backbone with two phosphoryl groups. The core polysaccharide region of V. cholerae contains keto-3-deoxy- D-mannose-octulosonic acid (KDO), D-glucose, heptose, D- fructose and ethanolamine phosphate. The O- polysaccharide side chain of V cholerae is a homo-polymer of D-perosamine approximately 17-18 units in length (Farmer et al., 2005). In the cell membrane of Vibrio spp., the major fatty acids are usually C16:1々7c/iso-C15:02-OH, C16:0, C18:1々7c and C14:0 (Wang et al., 2010; Wang et al., 2011). The major polar lipids are usually phosphatidylethanolamine and phosphatidylglycerol (Gao et al., 2012; Doi et al., 2016).
Many vibrios have been detected with capsules surrounding the cells. The carbohydrate composition of the capsule may vary from strain to strain; sugars include a-N- acetyl quinovosamine, a-N-acetyl galactosamine uronic acid, rhamnosamine, and fucosamine (Farmer et al., 2005).
When Vibrio spp. are infected by viruses or preyed upon by protozoans, some of the cellular carbohydrate composition will be released to the environment or be used as nutrient resources by protozoans for further biotransformation or remineralization processes.
4.2Carbon storage
Vibrio spp. may survive carbon starvation for a month or even longer, with carbon storage enabling growth in a fluctuating environment where individual resources may be limiting at various times (Thompson and Polz, 2006). In V. cholerae, carbon inclusion bodies, e.g., glycerol or poly-p- hydroxybutyrate (PHB), are formed in the presence of excess carbon (i.e., glucose), and are consumed in the case of carbon starvation (Thompson and Polz, 2006). 0vreas et al. (2003) reported that a large-celled V splendidus population filled with C-rich granules, i.e. PHB, became dominant when excess glucose along with inorganic (nitrate and phosphate) nutrient was added to seawater microcosms.
4.3Carbon products
Vibrios are able to engage in both respiratory and fermentative metabolism and transform organic carbon into cell material and the waste products of energy metabolism. During aerobic or anaerobic respiration, large amounts of metabolic end products are excreted (Farmer et al., 2005; Thompson and Polz, 2006). Common metabolic end products of D-glucose metabolism include organic acids (i.e., formic, acetic, lactic, succinic, and pyruvic acids), acetone, alcohols, CO2, and, in some cases(Farmer et al., 2005; Thompson and Polz, 2006).
5. Molecular mechanisms of Vibrio-mediated carbon cycling
5.1 Extracellular hydrolytic enzymes
Most dissolved and particulate organic matter in the marine environment is in the form of complex polymers that must be hydrolyzed to oligomers or monomers prior to cellular uptake. This is performed by extracellular hydrolytic enzymes with different substrate specificities. Vibrio spp. and other Gram-negative bacteria degrade complex organic matter through a defined sequence of steps. Partial hydrolysis of complex polymers must occur extracellularly prior to transport into the periplasmic space, where additional enzymes act to create monomers that can be transported into the cell cytoplasm (Thompson and Polz, 2006).
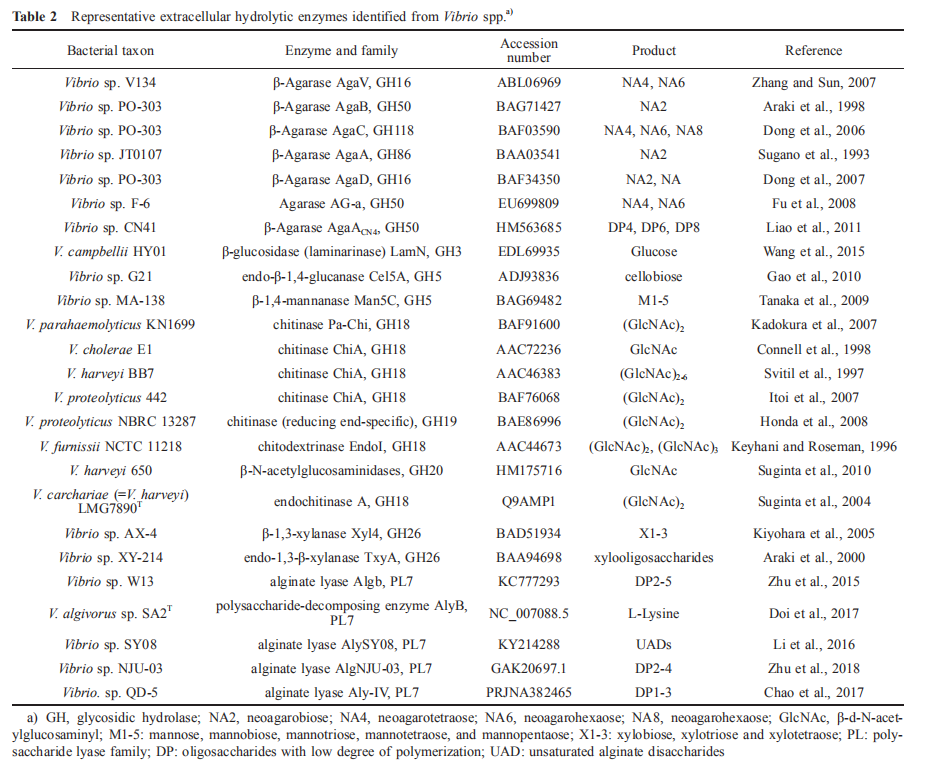
Various extracellular hydrolytic enzymes that can degrade polysaccharides, including chitinase, agarase, laminarinase and amylase, have been identified in Vibrio (Table 2). Chitinases, proteases, and lipases are among the cell surface or exuded hydrolases that have been described, whereas enzymatic activities, such as alkaline phosphatase and amino-peptidase, appear to be concentrated in the periplasmic space (Thompson and Polz, 2006).
5.2 Vibrio genomes
Whole genome sequences will greatly expand understanding of Vibrio biology, including ecology, and provide unprecedented opportunities to shed light on nutrient cycling in the ocean. The traditional phenotypic characterization of Vibrio has been expensive, time-consuming and restricted in scope to a limited number of features. A reliable and possible alternative is to obtain phenotypic information directly from whole genome sequences. Moreover, genomic information from whole genome sequences of Vibrio species has confirmed many of the previously published biological and ecological reports (Amaral et al., 2014).
Currently, there are >1500 genomes of Vibrio spp. available in the GenBank database, although complete genome sequences are available for only 19 Vibrio spp. (Lin et al., 2018). All the Vibrio spp. examined to date have two chromosomes. Chromosome 1 is usually larger, with a relatively constant size of ~3 Mb, encoding around 2700 proteins that represent many essential functions, such as ribosomal proteins, DNA replication machinery and the polar flagella. In contrast, chromosome 2 is smaller, ~1 Mb encoding approximately 1000 proteins, and contains few essential genes, but many undefined genes acquired by horizontal transfer, which may account for the fast replication rate and wide range of niche specialization of Vibrio spp. (Lukjancenko and Ussery, 2014; Lin et al., 2018).
A remarkable feature of all Vibrio spp. is of highly plastic genomes, which enables them to have a broad metabolic range, and use a wide variety of carbon sources. DOM provides much of the nutrition needed by Vibrio to live in estuarine and marine habitats. The chemical composition of DOM is highly variable. Neutral sugars account for 2% to 6% of the DOC in surface waters, and 0.3% to 0.9% in deep water (Benner, 2002). The most abundant hydrolysable neutral sugars are glucose, galactose, fucose, mannose, and xylose (Benner, 2002), and almost all of the genes encoding enzymes necessary to metabolize these common sugars are present in Vibrio genomes (Grimes et al., 2009). In addition, genes encoding enzymes degrade polysaccharides (such as chitin, cellulose) are present in Vibrio genomes (Grimes et al., 2009; Lin et al., 2018).

Marine microbial metagenomic technologies provide us with a descriptive and predictive metabolic and taxonomic model ofanecosystem (Gilbert and Dupont, 2011). Recently, the whole-genome enrichment approach has been used successfully for direct genotyping and metagenomic analysis of low abundant V. cholerae DNA from natural water (Vezzulli et al., 2017). It has been observed that Vibrio spp. could significantly alter carbon, nitrogen and other microbiome metabolism in specific niches, despite that metagenomic data revealed they might be in low abundance (Turner et al., 2009).
6. Conclusions and prospect
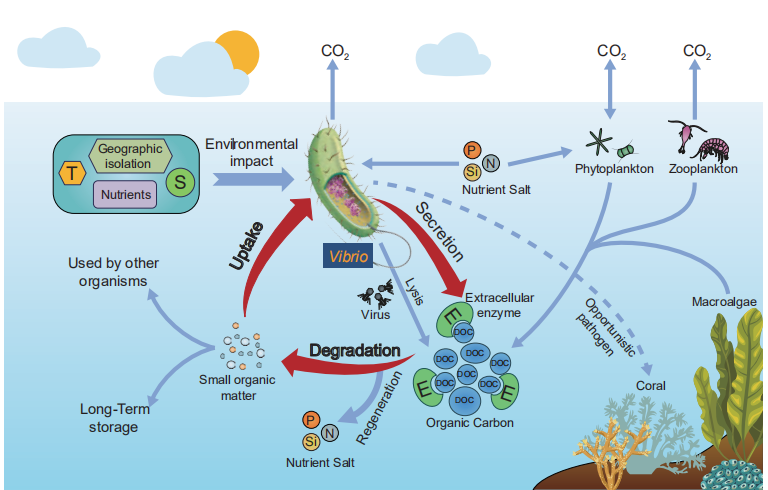
In summary, Vibrio spp. are widespread in the marine environment, and may consume a wide range of organic carbon sources. The extremely short replication time of Vibrio spp. may lead to extensive blooms, which occur in response to nutrient pulses. Vibrio spp. are undoubtedly key players in marine organic carbon cycles, especially in marginal seas. We propose a potential action mode of Vibrio species in marine organic carbon cycling (Figure 3). However, the exact contribution of Vibrio spp. to marine organic carbon cycles needs to be further investigated.
Firstly, although the abundance and community composition of Vibrio have been studied in many marine environments, some areas (e.g., Chinese marginal seas) are still under-investigated. Currently, most information regarding Vibrio ecology is derived from culturable vibrios. In fact, there are many uncultured Vibrio spp. in the environment, so both culture-based and culture-independent methods should be employed for future studies. In addition, as many Vibrio enter into the VBNC state under unfavorable environmental conditions, the activities of these cells need to be studied.

Figure 3 Potential action mode of Vibrio species in marine organic carbon cycling. T, temperature; S, salinity; P, phosphate; N, nitrogen salt; Si, silicate; E, extracellular enzyme; POC, particulate organic carbon; DOC, dissolved organic carbon.between Vibrio and viruses may well be an important process in carbon cycling, but the detailed contribution and mechanisms needs to be further elucidated.
Secondly, it has been proved that DOC, in addition to temperature and salinity, has a strong effect on the distribution of Vibrio. However, the effect of DOC is multifactorial, as the bulk DOC has different organic components, which may impact Vibrio growth differentially. Clearly, analysis of the chemical composition (such as glucose, chitin and alginate) of DOC and the correlation with the distribution of Vibrio population will give more information regarding the ecology of Vibrio spp. In addition, the interaction Nutrient Salt
Thirdly, it has been demonstrated that Vibrio are able to attack many organic carbon compounds. However, the range of organic carbon substrates that may be used by Vibrio has been determined largely from conventional phenotypic testing. The extent of organic carbon substrates, especially from various phytoplankton and zooplankton polysaccharides, should be thoroughly investigated in the future.
Finally, the interactions between Vibrio population and organic matter are crucial for the biogeochemical cycles in marine system. Currently, only a few details exist regarding the molecular components and mechanisms involved in Vibrio-mediated biotransformation and remineralization processes. More efforts need to be conducted to elucidate the genetic basis of these processes. A better understanding is needed of what molecular machinery is present in Vibrio that allows the cells to best utilize the diverse variety of DOM found in marine and estuarine habitats. Whole genome sequences have refined our understanding of adaptations, physiology and ecology of the genus Vibrio, and the advent of the metagenomic data (especially useful for uncultured Vibrio spp.) will further elucidate their roles on organic carbon cycling in the ocean.
Acknowledgements This work was supported by the National Natural Science Foundation of China (Grant Nos. 41730530, 91751202, 41476112) and the National Key Research and Development Program of China (Grant No. 2016YFA0601303).
References
Amaral G R S, Dias G M, Wellington-Oguri M, Chimetto L, Campeao M E, Thompson F L, Thompson C C. 2014. Genotype to phenotype: Identification of diagnostic vibrio phenotypes using whole genome sequences. Int J Syst Evol Microbiol, 64: 357-365
Amin A K M R, Feng G, Al-saari N, Meirelles P M, Yamazaki Y, Mino S, Thompson F L, Sawabe T, Sawabe T. 2016. The first temporal and spatial assessment of vibrio diversity of the surrounding seawater of coral reefs in ishigaki, Japan. Front Microbiol, 7: 1185
Araki T, Hayakawa M, Lu Z, Karita S, Morishita T. 1998. Purification and characterization of agarases from a marine bacterium, Vibrio sp. PO- 303. J Mar Biotechnol, 6: 260-265
Araki T, Hashikawa S, Morishita T. 2000. Cloning, sequencing, and expression in Escherichia coli of the new gene encoding beta -1,3-Xy- lanase from a marine bacterium, Vibrio sp. strain XY-214. Appl Environ Microbiol, 66: 1741-1743
Arnosti C. 2014. Patterns of microbially driven carbon cycling in the ocean: Links between extracellular enzymes and microbial communities. Adv Oceanogr, 2014: 1-12
Austin B, Zhang X H. 2006. Vibrio harveyi: A significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol, 43: 119-124 Baker-Austin C, Trinanes J, Gonzalez-Escalona N, Martinez-Urtaza J.
2017. Non-cholera Vibrios: The microbial barometer of climate change. Trends Microbiol, 25: 76-84
Azam F, Malfatti F. 2007. Microbial structuring of marine ecosystems. Nat Rev Microbiol, 5: 782-791
Beijerinck M W. 1889. Le Photobacterium luminosum, bacterie lumineuse de la Mer du Nord. Archives Neerlandaises des Sciences Exactes et Naturelles, 23: 401-427
Benner R. 2002. Chemical composition and reactivity. In: Hansell D A, Carlson C A, eds. Biogeochemistry of Marine Dissolved Organic
Matter. New York : Academic. 59-90
Bruhn J B, Nielsen K F, Hjelm M, Hansen M, Bresciani J, Schulz S, Gram L. 2005. Ecology, inhibitory activity, and morphogenesis of a marine antagonistic bacterium belonging to the Roseobacter Clade. Appl Environ Microbiol, 71: 7263-7270
Chan K, Woo M, Lo K, French G. 1986. Occurrence and distribution of halophilic vibrios in subtropical coastal waters of Hong Kong. Appl Environ Microbiol, 52: 1407-1411
Chao Y, Wang S, Wu S, Wei J, Chen H. 2017. Cloning and characterization of an alginate lyase from marine Vibrio. sp. QD-5. Preprints
Chi W J, Chang Y K, Hong S K. 2012. Agar degradation by microorganisms and agar-degrading enzymes. Appl Microbiol Biotechnol, 94: 917-930
Chimetto Tonon L A, Thompson J R, Moreira APB, Garcia G D, Penn K, Lim R, Berlinck R G S, Thompson C C, Thompson F L. 2017. Quantitative detection of active vibrios associated with white plague disease inMussismilia braziliensis corals. Front Microbiol, 8: 2272
Connell T D, Metzger D J, Lynch J, Folster J P. 1998. Endochitinase is transported to the extracellular milieu by the eps-encoded general secretory pathway of Vibrio cholerae. J Bacteriol, 180: 5591-5600
Davis B J K, Jacobs J M, Davis M F, Schwab K J, DePaola A, Curriero F C. 2017. Environmental determinants of Vibrio parahaemolyticus in the Chesapeake Bay. Appl Environ Microbiol, 83: e01147-17
Doi H, Chinen A, Fukuda H, Usuda Y. 2016. Vibrio algivorus sp. nov., an alginate- and agarose-assimilating bacterium isolated from the gut flora of a turban shell marine snail. Int J Syst Evol Microbiol, 1: 3164-3169
Doi H, Tokura Y, Mori Y, Mori K, Asakura Y, Usuda Y, Fukuda H, Chinen
A.2017. Identification of enzymes responsible for extracellular alginate depolymerization and alginate metabolism in Vibrio algivorus. Appl Microbiol Biotechnol, 101: 1581-1592
Dong J, Hashikawa S, Konishi T, Tamaru Y, Araki T. 2006. Cloning of the novel gene encoding-agarase C from a marine bacterium, Vibrio sp. strain PO-303, and characterization of the gene product. Appl Environ Microbiol, 72: 6399-6401
Dong J, Tamaru Y, Araki T. 2007. Molecular cloning, expression, and characterization of a p-agarase gene, agaD, from a marine bacterium, Vibrio sp. strain PO-303. Biosci Biotech Biochem, 71: 38-46
Eilers H, Pernthaler J, Glockner F O, Amann R. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol, 66: 3044-3051
Eiler A, Johansson M, Bertilsson S. 2006. Environmental influences on Vibrio populations in northern temperate and boreal coastal waters (Baltic and Skagerrak Seas). Appl Environ Microbiol, 72: 6004~6011
Farmer J J, Janda J M, Brenner F W, Cameron D N, Birkhead K M. 2005. Genus I. Vibrio Pacini 1854. In: Brenner D J, Kreig N R, Staley J T, eds. Bergey's Manual of Systematic Bacteriology. 2nd ed. New York: Springer Science Business Media. 494-546
Feller G, Narinx E, Arpigny J L, Zekhnini Z, Swings J, Gerday C. 1994. Temperature dependence of growth, enzyme secretion and activity of psychrophilic Antarctic bacteria. Appl Microbiol Biotechnol, 41: 477479
Fu W, Han B, Duan D, Liu W, Wang C. 2008. Purification and characterization of agarases from a marine bacterium Vibrio sp. F-6. J Ind Microbiol Biotechnol, 35: 915-922
Fujino T, Okuno Y, Nakada D, Aoyama A, Fukai K, Mukai T, Ueha T. 1951. On the bacteriological examination of shirasu food poisoning (in Japanese). J Jpn Assoc Inf Dis, 25: 11
Gao Z, Ruan L, Chen X, Zhang Y, Xu X. 2010. A novel salt-tolerant endo- p-1,4-glucanase Cel5A in Vibrio sp. G21 isolated from mangrove soil. Appl Microbiol Biotechnol, 87: 1373-1382
Gao Z M, Xiao J, Wang X N, Ruan L W, Chen X L, Zhang Y Z. 2012. Vibrio xiamenensis sp. nov., a cellulase-producing bacterium isolated from mangrove soil. Int J Syst Evol Microbiol, 62: 1958-1962
Gilbert J A, Dupont C L. 2011. Microbial metagenomics: Beyond the genome. Annu Rev Mar Sci, 3: 347-371
Gilbert J A, Steele J A, Caporaso J G, Steinbruck L, Reeder J, Temperton
B,Huse S, McHardy A C, Knight R, Joint I, Somerfield P, Fuhrman J A, Field D. 2012. Defining seasonal marine microbial community dynamics. ISME J, 6: 298-308
Girard L, Peuchet S, Servais P, Henry A, Charni-Ben-Tabassi N, Baudart J. 2017. Spatiotemporal dynamics of total viable Vibrio spp. in a NW Mediterranean coastal area. Microbes Environ, 32: 210-218
Gomez-Gil B, Thompson C C, Matsumura Y, Sawabe T, Iida T, Christen R. 2014. Family Vibrionaceae (Chapter 225). In: Rosenberg E, DeLong E, Thompson F L, Lory S, Stackebrandt E, eds. The Prokaryotes. 4th ed. New York: Springer. 88
Grimes D J, Johnson C N, Dillon K S, Flowers A R, Noriea N F, Berutti T. 2009. What genomic sequence information has revealed about Vibrio ecology in the ocean一A Review. Microb Ecol, 58: 447X60
Guerinot M L, West P A, Lee J V, Colwell R R. 1982. Vibrio diazotrophicus sp. nov., a Marine Nitrogen-Fixing Bacterium. Int J Systatic Bacteriology, 32: 350-357
Hamdan L, Fulmer P. 2011. Effects of COREXIT? EC9500A on bacteria from a beach oiled by the Deepwater Horizon spill. Aquat Microb Ecol, 63: 101-109
Hickey M E, Lee J L. 2017. A comprehensive review of Vibrio (Listonella) anguillarum: Ecology, pathology and prevention. Rev Aquacult, 161
Honda Y, Taniguchi H, Kitaoka M. 2008. A reducing-end-acting chitinase from Vibrio proteolyticus belonging to glycoside hydrolase family 19. Appl Microbiol Biotechnol, 78: 627-634
Itoi S, Kanomata Y, Koyama Y, Kadokura K, Uchida S, Nishio T, Oku T, Sugita H. 2007. Identification of a novel endochitinase from a marine bacterium Vibrio proteolyticus strain No. 442. BBA-Proteins Proteom, 1774: 1099-1107
Joseph S W, Colwell R R, Kaper J B. 1982. Vibrio Parahaemolyticus and related halophilic vibrios. Crit Rev Microbiol, 10: 77-124
Kadokura K, Rokutani A, Yamamoto M, Ikegami T, Sugita H, Itoi S, Hakamata W, Oku T, Nishio T. 2007. Purification and characterization of Vibrio parahaemolyticus extracellular chitinase and chitin oligosaccharide deacetylase involved in the production of heterodisaccharide from chitin. Appl Microbiol Biotechnol, 75: 357-365
Kauffman K M, Hussain F A, Yang J, Arevalo P, Brown J M, Chang W K, VanInsberghe D, Elsherbini J, Sharma R S, Cutler M B, Kelly L, Polz M F. 2018. A major lineage of non-tailed dsDNA viruses as unrecognized killers of marine bacteria. Nature, 554: 118-122
Keyhani N O, Roseman S. 1996. The chitin catabolic cascade in the Marine Bacterium Vibrio fumissii. J Biol Chem, 271: 33414-33424
Kirchman D, White J. 1999. Hydrolysis and mineralization of chitin in the Delaware Estuary. Aquat Microb Ecol, 18: 187-196
Kiyohara M, Sakaguchi K, Yamaguchi K, Araki T, Nakamura T, Ito M. 2005. Molecular cloning and characterization of a novel p-1,3-xylanase possessing two putative carbohydrate-binding modules from a marine bacterium Vibrio sp. strain AX-4. Biochem J, 388: 949-957
Kopprio G A, Streitenberger M E, Okuno K, Baldini M, Biancalana F, Fricke A, Martinez A, Neogi S B, Koch B P, Yamasaki S, Lara R J. 2017. Biogeochemical and hydrological drivers of the dynamics of Vibrio species in two Patagonian estuaries. Sci Total Environ, 579: 646656
Lapota D, Galt C, Losee J R, Huddell H D, Orzech J K, Nealson K H. 1988. Observations and measurements of planktonic bioluminescence in and around a milky sea. J Exp Mar Biol Ecol, 119: 55-81
Lehmann K B, Neumann R. 1896. Atlas und Grundriss der Bakteriologie und Lehrbuch der speziellen bakteriologischen Diagnostik. 1st ed. J F Lehmann, Munchen
Li X, Roseman S. 2004. The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/ kinase. Proc Natl Acad Sci USA, 101: 627-631
Li S, Wang L, Hao J, Xing M, Sun J, Sun M. 2016. Purification and characterization of a new alginate lyase from marine bacterium Vibrio sp. SY08. Mar Drugs, 15: 1
Liao L, Xu X W, Jiang X W, Cao Y, Yi N, Huo Y Y, Wu Y H, Zhu X F, Zhang X Q, Wu M. 2011. Cloning, expression, and characterization of a new p-Agarase from Vibrio sp. strain CN41. Appl Environ Microbiol, 77: 7077-7079
Lin H, Yu M, Wang X, Zhang X H. 2018. Comparative genomic analysis reveals the evolution and environmental adaptation strategies of vibrios. BMC Genomics, 19: 135
Liu Z, Liu J. 2013. Evaluating bacterial community structures in oil collected from the sea surface and sediment in the northern Gulf ofMexico after the Deepwater Horizon oil spill. Microbiol Open, 2: 492-504
Lukjancenko O, Ussery D W. 2014. Vibrio chromosome-specific families. Front Microbiol, 5: 73
Main C R, Salvitti L R, Whereat E B, Coyne K J. 2015. Community-level and species-specific associations between phytoplankton and particle- associated Vibrio species in Delaware's Inland Bays. Appl Environ Microbiol, 81: 5703-5713
Martinez A, Ventouras L A, Wilson S T, Karl D M, DeLong E F. 2013. Metatranscriptomic and functional metagenomic analysis of methylphosphonate utilization by marine bacteria. Front Microbiol, 4
Machado H, Gram L. 2015. The fur gene as a new phylogenetic marker for Vibrionaceae species identification. Appl Environ Microbiol, 81: 2745- 2752
Miller S D, Haddock S H D, Elvidge C D, Lee T F. 2005. Detection of a bioluminescent milky sea from space. Proc Natl Acad Sci USA, 102: 14181-14184
Oberbeckmann S, Fuchs B M, Meiners M, Wichels A, Wiltshire K H, Gerdts G. 2012. Seasonal dynamics and modeling of a Vibrio community in coastal waters of the North Sea. Microb Ecol, 63: 543-551
Oliver J D. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. Fems Microbiol Rev, 34: 415-425
Ovreas L, Bourne D, Sandaa R, Casamayor E, Benlloch S, Goddard V, Smerdon G, Heldal M, Thingstad T. 2003. Response of bacterial and viral communities to nutrient manipulations in seawater mesocosms. Aquat Microb Ecol, 31: 109-121
Pascual J, Macian M C, Arahal D R, Garay E, Pujalte M J. 2010. Multilocus sequence analysis of the central clade of the genus Vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int J Syst Evol Microbiol, 60: 154-165
Phillips K E, Satchell K J F. 2017. Vibrio vulnificus: From oyster colonist to human pathogen. PLoS Pathog, 13: e1006053
Rizzo L, Fraschetti S, Alifano P, Tredici M S, Stabili L. 2016. Association of Vibrio community with the Atlantic Mediterranean invasive alga Caulerpa cylindracea. J Exp Mar Biol Ecol, 475: 129-136
Rubio-Portillo E, Gago J F, Martinez-Garcia M, Vezzulli L, Rossello-Mora R, Anton J, Ramos-Espla A A. 2018. Vibrio communities in scler- actinian corals differ according to health status and geographic location in the Mediterranean Sea. Syst Appl Microbiol, 41: 131-138
Rubio-Portillo E, Yarza P, Penalver C, Ramos-Espla A A, Anton J. 2014. New insights into Oculina patagonica coral diseases and their associated Vibrio spp. communities. ISME J, 8: 1794-1807
Ruby E G, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg E P. 2005. Complete genome sequence of Vibrio fischeri: A symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci USA, 102: 3004-3009
Sakazaki R, Iwanami S, Fukumi H. 1963. Studies on the enteropathogenic facultatively halophilic bacteria Vibrio parahaemolyticus. I. Morphological, cultural and biochemical properties and its taxonomical position. Jpn J Med Sci Biol, 16: 161-188
Siboni N, Balaraju V, Carney R, Labbate M, Seymour J R. 2016. Spatiotemporal dynamics of Vibrio spp. within the Sydney harbour estuary. Front Microbiol, 7: 460
Simu K, Hagstrom A. 2004. Oligotrophic bacterioplankton with a novel single-cell life strategy. Appl Environ Microbiol, 70: 2445-2451
Sneha K G, Anas A, Jayalakshmy K V, Jasmin C, Das P V V, Pai S S, Pappu S, Nair M, Muraleedharan K R, Sudheesh K, Nair S. 2016. Distribution of multiple antibiotic resistant Vibrio spp. across Palk Bay. Region Stud Mar Sci, 3: 242-250
Sugano Y, Matsumoto T, Kodama H, Noma M. 1993. Cloning and sequencing of agaA, a unique agarose 0107 gene from a marine bacterium, Vibrio sp. strain JT0107. Appl Environ Microbiol, 59: 3750-3756 Suginta W, Chuenark D, Mizuhara M, Fukamizo T. 2010. Novel p-N- acetylglucosaminidases from Vibrio harveyi 650: Cloning, expression, enzymatic properties, and subsite identification. BMC Biochem, 11: 40 Suginta W, Vongsuwan A, Songsiriritthigul C, Prinz H, Estibeiro P, Duncan R R, Svasti J, Fothergill-Gilmore L A. 2004. An endochitinase A from Vibrio carchariae: Cloning, expression, mass and sequence analyses, and chitin hydrolysis. Archives Biochem Biophys, 424: 171-180
Svitil A L, Chadhain S, Moore J A, Kirchman D L. 1997. Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl Environ Microbiol, 63: 408413
Takemura A F, Chien D M, Polz M F. 2014. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol, 5: 38
Tall A, Hervio-Heath D, Teillon A, Boisset-Helbert C, Delesmont R, Bodilis J, Touron-Bodilis A. 2013. Diversity of Vibrio spp. isolated at ambient environmental temperature in the Eastern English Channel as determined by pyrH sequencing. J Appl Microbiol, 114: 1713-1724
Tanaka M, Umemoto Y, Okamura H, Nakano D, Tamaru Y, Araki T. 2009. Cloning and characterization of a p-1,4-mannanase 5C possessing a family 27 carbohydrate-binding module from a marine bacterium,Vibrio sp. strain MA-138. Biosci Biotech Biochem, 73: 109-116
Thompson F L, Austin B, Swings J. 2006. The Biology of Vibrios. Washington D C: American Society for Microbiology
Thompson F L, Gevers D, Thompson C C, Dawyndt P, Naser S, Hoste B, Munn C B, Swings J. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol, 71: 5107-5115
Thompson J R, Polz M F. 2006. Dynamics of Vibrio populations and their role in environmental nutrient cycling. In: Thompson F L, Austin B, Swings J, eds. The Biology of Vibrios. Washington D C: ASM Press. 190-203
Thompson J R, Randa M A, Marcelino L A, Tomita-Mitchell A, Lim E, Polz M F. 2004. Diversity and dynamics of a north atlantic coastal Vibrio community. Appl Environ Microbiol, 70: 4103-4110
Turner J W, Good B, Cole D, Lipp E K. 2009. Plankton composition and environmental factors contribute to Vibrio seasonality. ISME J, 3: 1082-1092
Thurber R V, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards R A, Angly F, Dinsdale E, Kelly L, Rohwer F. 2009. Metagenomic analysis of stressed coral holobionts. Environ Microbiol, 11: 2148-2163
Vezzulli L, Brettar I, Pezzati E, Reid P C, Colwell R R, Hofle M G, Pruzzo
C.2012. Long-term effects of ocean warming on the prokaryotic community: Evidence from the vibrios. ISME J, 6: 21-30
Vezzulli L, Grande C, Reid P C, Helaouet P, Edwards M, Hofle M G, Brettar I, Colwell R R, Pruzzo C. 2016. Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc Natl Acad Sci USA, 113: E5062-E5071
Vezzulli L, Grande C, Tassistro G, Brettar I, Hofle M G, Pereira R P A, Mushi D, Pallavicini A, Vassallo P, Pruzzo C. 2017. Whole-genome enrichment provides deep insights into Vibrio cholerae Metagenome from an African River. Microb Ecol, 73: 734-738
Vezzulli L, Pezzati E, Moreno M, Fabiano M, Pane L, Pruzzo C, Pruzzo C. 2009. Benthic ecology of Vibrio spp. and pathogenic Vibrio species in a coastal Mediterranean environment (La Spezia Gulf, Italy). Microb Ecol, 58: 808-818
Wang H, Liu J, Wang Y, Zhang X H. 2011. Vibrio marisflavi sp. nov., a novel marine bacterium isolated from seawater near the Yellow Sea Cold Water Mass, China. Int J Syst Evol Microbiol, 61: 568-573
Wang Y, Zhang X H, Yu M, Wang H, Austin B. 2010. Vibrio atypicus sp. nov., isolated from the digestive tract of the Chinese prawn (Penaeus chinensis O'sbeck). Int J Syst Evol Microbiol, 60: 2517-2523
Wang Z, Robertson K L, Liu C, Liu J L, Johnson B J, Leary D H, Compton J R, Vuddhakul V, Legler P M, Vora G J. 2015. A novel Vibrio betaglucosidase (LamN) that hydrolyzes the algal storage polysaccharide laminarin. Fems Microbiol Ecol, 91: fiv087
West P A, Okpokwasili G C, Brayton P R, Grimes D J, Colwell R R. 1984. Numerical taxonomy of phenanthrene-degrading bacteria isolated from the Chesapeake Bay. Appl Environ Microbiol, 48: 988-993
Westrich J R, Ebling A M, Landing W M, Joyner J L, Kemp K M, Griffin D W, Lipp E K. 2016. Saharan dust nutrients promote Vibrio bloom formation in marine surface waters. Proc Natl Acad Sci USA, 113: 59645969
Xu H S, Roberts N, Singleton F L, Attwell RW, Grimes D J, Colwell R R. 1982. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol, 8: 313-323
Zhang W, Sun L. 2007. Cloning, characterization, and molecular application of a beta-agarase gene from Vibrio sp. strain V134. Appl Environ Microbiol, 73: 2825-2831
Zhu B, Tan H, Qin Y, Xu Q, Du Y, Yin H. 2015. Characterization of a new endo-type alginate lyase from Vibrio sp. W13. Int J Biol Macromol, 75: 330-337
Zhu B, Sun Y, Ni F, Ning L, Yao Z. 2018. Characterization of a new endotype alginate lyase from Vibrio sp. NJU-03. Int J Biol Macromol, 108: 1140-1147
(Responsible editor: Rui ZHANG)

