Carol Robinson1,*, Douglas Wallace2, Jung-Ho Hyun3, Luca Polimene4, Ronald Benner5, Yao Zhang6, Ruanhong Cai6, Rui Zhang6 and Nianzhi Jiao6
INTRODUCTION
The persistenceof the vast pool of recalcitrant dissolved organic carbon (RDOC) in the deep ocean is fundamental to both global carbon cycling and global climate. Yet, quantitative understanding of the mechanisms that produce and utilize RDOC is still in its infancy. Moving from a conceptual framework to quantitative understanding in marine carbon cycling can take the international research community several decades一precious time that is not available in this era of anthropogenic climate change. This perspective paper sets out an implementation strategy to move efficiently from conceptual hypotheses of marine carbon storage to quantification of the capacity for marine carbon storage through the microbial carbon pump (MCP), its sensitivity to global change and potential for modification. The aim is to excite and facilitate the international scientific community to hasten this process via an internationally co-ordinated multidisciplinary research initiative.
The marine microbial carbon pump (MCP) [1] produces a concentration gradient of reactivity in dissolved organic carbon (DOC) from low concentrations of reactive fractions of DOC that have an average lifetime of less than 100 years (DOC<ioo) to high concentrations of recalcitrant DOC (RDOC) fractions that persist for longer than 100 years (DOC>ioo) [2]. This gradient is maintained against the continuous degradation of DOC>i00 by photochemical transformation in surface waters, elevated temperatures at deep hydrothermal vents and gradual microbial degradation throughout the water column [1-3]. The resultant pool of RDOC amounts to the sequestration of carbon in the ocean equivalent to the amount of carbon dioxide in the atmosphere. The magnitude of the MCP is determined as the rate ofpro- duction of DOC>100 in mol C m-2 y-1 [2]. Global warming, weakening thermohaline circulation, increasing UV radiation, ocean acidification, increasing oxygen minimum zones and increasing nutrient concentrations are all processes potentially affecting the magnitude of the MCP, leading to increased or decreased transformation of marine RDOC to atmospheric CO2 with associated consequences for global climate [3,4].
In order to quantify the MCP, and understand its sensitivity to large-scale environmental change, the rates of production and loss of RDOC via each potential formation and loss pathway need to be determined and understood. This is a major challenge, likely beyond the ability of any individual team of researchers, given the complexity and diversity of the substances, processes and microbial organisms involved and the variety of controlling factors. However, the connectivity and biogeographical characterization of the ocean一a consequence of ocean circulation and mixing一allow a co-ordinated approach where results from multiple groups, working at different locations and times, could be compared and assembled in order to achieve progress.
Such a co-ordinated approach will require a multi-component programme of systematic experimentation linked with related conceptual and numerical modelling. The suggested programme components include: (i) determination of the chemical components of RDOC, including ‘fingerprint' constituents to identify the presence of RDOC; (ii) a standardized and inter compa rable programme of long-term microcosm incubation experiments conducted worldwide to determine loss of labile DOC, and net accumulation or loss of bulk RDOC, under specific environmental conditions; (iii) the concurrent determination and quantification of RDOC production and loss pathways; and (iv) quantification of the presence and activity of the responsible microbial organisms and their genetic potential. In order to test our understanding as well as the efficacy of our experimental protocols, a linked ‘backbone' programme of larger-scale, controlled manipulation experiments and related, long-term time-series studies should be established (v). This should involve controllable meso- and macro-cosm facilities as well as in-situ time-series studies where the determination of rate processes, RDOC composition change (and associated proxies), microbial community composition and genetic potential can be examined repeatedly, under varying conditions. The programme needs to be structured within (vi) a framework of conceptual and numerical models and, crucially, in order to assess the global context of RDOC production and loss, (vii) large spatial- scale and long temporal-scale surveys of, at minimum, the fingerprints or proxies of RDOC accumulation will be required. The surveys will allow experimental findings to be parameterized in global biogeochemical models to explore the sensitivity of the MCP and associated carbon storage to environmental change over long timescales.
For each of these programme components, we describe here the principle of the approach, current limitations and future research requirements. This perspective paper will therefore provide a route by which to progress from the conceptual framework of the MCP, to the quantification of the microbial processes that produce and utilize RDOC, and the environmental, trophic and physiological influences on them.
Identification of the components of RDOC
The very nature of RDOC, in terms of its complexity, diversity, low concentration and longevity, means that identifying representative components of RDOC is a major analytical challenge.
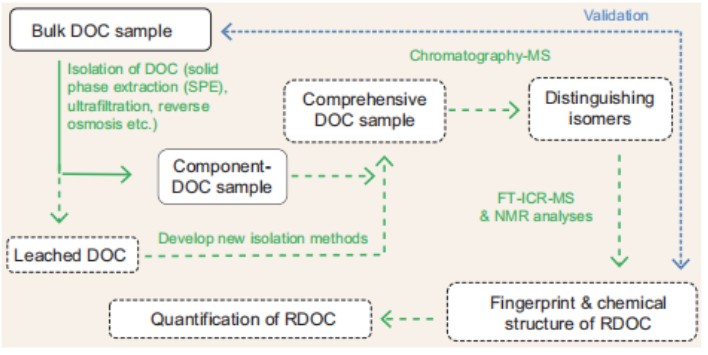
A variety of methods are now available to extract low-molecular-weight, 14C-depleted RDOC and nuclear magnetic resonance (NMR) spectrometry, gas chromatography-mass spectrometry (GC-MS) and Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR-MS) can identify specific chemical fingerprints of this material, including carboxyl-rich alicyclic molecules (C^AM), material derived from linear terpenoids and a group of stable molecular formulae known as the ‘island of stability'.
However, the comprehensive analysis of DOC and quantification of RDOC are still far from complete. Current techniques are limited in that existing isolation methods including solid phase extraction (SPE) can only isolate representative fractions of DOC, some of which are further discriminated during mass spectrometry analysis. In addition, direct injection of the DOC sample into the mass spectrometer cannot differentiate the numerous possibilities of isomer configuration for an assigned molecular formula, which therefore precludes the identification of an unambiguous chemical structure [5].
Future approaches should include the use of combined methods for DOC isolation and the development of new techniques to isolate the DOC (Fig. 1) [6]. Chromatographic analysis prior to ultra- high-resolution mass spectrometry could help to differentiate isomers, and subsequent analyses by a combination of FT- ICR-MS and high-field NMR would allow progress towards the ultimate goal of revealing the structures and concentration of representative RDOC molecules.
Incubation experiments to determine loss of labile DOC and accumulation of RDOC
Operationally, the reactivity of DOC is determined from the loss of DOC over a given period of time [7], with a biologically labile fraction (LDOC) defined as that which is degraded over hours to days, a semi-labile fraction (SLDOC) that is removed in weeks to months and a semi-refractory fraction (SRDOC) with turnover times of years to decades. Studies of the degradation of LDOC involve the incubation of seawater samples under controlled laboratory conditions for days to years, allowing the calculation of the DOC decay constant kc over time [8].
However, there is no standard method by which to undertake these experiments and, since both the concentration and decay rate of DOC are dependent on time and incubation conditions such as temperature and nutrients, collation and comparison of the available dataare problematic. Methodological differences include the method by which a diluted seawater sample is obtained, such as the composition and pore size of the filters used, the ratio of filtrate to inoculum, the incubation temperature, light and nutrient conditions, and the volume and time period of the incubation.
A comparison of methods and recommendations for standardization are required to advance this aspect of DOC reactivity research, following the approach taken recently by the research community investigating the impact of ocean acidification on marine plankton activity [9]. Once standardized methods are defined, systematic laboratory-scale bioassay experiments can be undertaken to assess the influence of environmental conditions such as temperature, light and nutrients on DOC reactivity, and incorporation of measurements of RDOC components or proxies of RDOC will allow the accumulation of RDOC to be determined alongside the loss of DOC.
Figure 1. Schematic of an experimental procedure to chemically characterize RDOC. The solid linesand boxes indicate well-established analyses, while the dashed lines and boxes indicate those thatare still to be explored.
Identification and quantification of RDOC production and loss pathways
The ocean reservoir of RDOC is determined by the net effect of its production and removal processes. Therefore, it is essential to investigate the mechanisms of both production and loss pathways in order to understand the influence of global climate change on ocean carbon sequestration.
Potential RDOC production pathways include phytoplankton excretion and autolysis, viral lysis, bacterial and ar- chaeal transformation, and protistan and zooplankton grazing andexcretion. A systematic approach is needed to determine the rate of production of ‘model' RDOC compounds from each of these pathways. For example, a range of RDOC biomarker compounds that are only produced by heterotrophic bacteria, including muramic acid and D-enantiomers of amino acids, have been identified. Quantitative measurements of these bacterial biomarkers in seawater-dissolved organic matter indicate that about 25% of the carbon and 50% of the nitrogen is of bacterial origin [10]. Based on biomarker data and bioassay experiments, the rate of production of bacterial RDOC in the upper 200 m of the ocean is estimated to be 8-23 TgC year-1 [11].
The persistence of RDOC in the ocean is well known, but the reasons for this persistence are still being explored [4,12]. Three broad areas of research are being investigated: (i) intrinsic properties of molecules, such as chemical composition and structure; (ii) environmental conditions, such as microbial community structure and exposure to solar radiation; and (iii) concentrations of molecules that are below a critical threshold for microbial utilization.Multiple and independent approaches are needed to delve deeper into the mysteries of RDOC removal from the ocean. For example, recent bioassay experiments have provided evidence that some RDOC is too diluted for microbial utilization [13] and that intrinsic properties of RDOC and environmental conditions limit its microbial utilization [3]. A modelling approach addressing these same questions provided independent evidence indicating the dilution of labile substrates likely accounts for a relatively small fraction of the RDOC in the deep ocean [14]. It appears that most RDOC persists in the deep ocean due to intrinsic properties and environmental conditions. Ocean mixing moves RDOC into different environmental conditions, and exposure to solar radiation and different microbial communities plays a critical role in its removal [3].
Stable isotope probing (SIP) [15] can be used to ascertain which organisms are responsible for DOC degradation. SIP tracks the incorporation of isotopically enriched substrates into the DNA, RNA or phospholipid fatty acids of these organisms. Combined with high- throughput sequencing, DNA/RNA SIP can identify which organisms are responsible for any observed biodegradation pathway with high phylogenetic resolution. Thus, incubation experiments with additions of13C-labelled labile DOC and subsequent 13C- and 12C-DNA analysis with terminal-restriction fragment length polymorphism (T-RFLP) and pyrosequencing can identify the microbial populations that incorporate the 13C- labelled substrate. The molecular characteristics of the produced 13C-labelled DOC (relatively more refractory) can be further determined using FT-ICR-MS, NMR or excitation emission matrix (EEM) fluorescence. Unfortunately, SIP techniques are not inherently quantitative. Substrate assimilation rates can be derived from changes in 513POC (particulate organic carbon) during concurrent incubations to help link the DOC-specific degradation rates to the activity of the microbial population.
Future work should use laboratory- controlled phytoplankton, zooplankton and bacterial cultures to produce DOC from each potential RDOC formation pathway to identify potential chemical biomarkers for each. The recent connection between RDOC fractions and precursors such as phytoplankton carotenoids lays the foundation for hypothesis testing of these potential transformation pathways. Long-term degradation incubations incorporating DNA/RNA SIP couldthenbe conducted to assess the bioreactivity of DOC from each of these formation pathways.
Microbial community composition and genetic potential
The microbial transformation of shortlived particulate and dissolved organic carbon (POC<1oo and DOC<1oo) to DOC>1oo is ultimately driven by the activity of microbial functional genes [16]. For example, genes for the ATP-binding cassette (ABC) transporter, including importers and exporters, are thought to be indicators of the capabilities of microbes to use or generate corresponding DOC compounds [16]. Therefore, a strategy to quantify the various processes of the MCP could depend on the quantification of microbial functional genes.
The quantification methods (such as PCR, qPCR, omics) usually used in ecological studies are time-consuming and potentially incomplete when considering the extremely high diversity of microbial functional genes involved in MCP processes. Microarrays that contain numerous probes targeting hundreds or thousands of genes are a promising technique in this regard. The development ofafunc- tional gene microarray involves gene selection and sequence retrieval from a database, probe design and evaluation, and microarray fabrication.
GeoChip is one of the most comprehensive functional gene arrays for quantification of the functional diversity, metabolic potential/activity and dynamics of microbial communities [17]. GeoChip 4 contains ~82000 probes covering > 140 000 coding sequences from 410 functional gene families, many of which are encoding biogeochemically important functions, such as carbon fixation and carbon degradation. In addition, GeoChip 4 has the ability to analyse targeted functional gene families of microorganisms in all four domains (Bacteria, Archaea, eukaryotes and viruses). A recent case study performed in the East China Sea suggested a possible connection between GeoChip data and MCP processes such as metabolic preferences of microbes along an environmental gradient and the degradation of recalcitrant organic matter (e.g. aromatic carboxylic acid and chlorinated aromatics) at depth [18].
However, a major technical limitation of GeoChip and other functional gene microarrays is the inability to interrogate novel sequences and, therefore, find novel biogeochemical pathways. Combination with high-throughput metage- nomic analysis may be an option. In the last decade, with the rapid progress in sequencing techniques, microbiologists have accumulated an unprecedented amount of genetic data (e.g. Tara Oceans Expedition (https://oceans. taraexpeditions.org/en/m/about-tara/), Global Ocean Sampling Expedition (GOS; http://www.jcvi.org/ cms/ research/projects/gos/overview/) and provided fundamental new understanding of biogeochemical cycles. This required the inclusion of probes for newly recognized functional genes and rapid updates of the microarray.
Assigning an exact function to a particular gene is a slow process, and significantly impedes the linkage between genetic sequence data and DOC transformation pathways [19]. Nevertheless, progress is being made. Recent studies have directly linked geochemical dynamics to the genetic composition of microbial communities [20] and the Tara Ocean microbial genomic data were successfully connected to carbon export in the oligotrophic ocean, which could even be predicted from a few bacterial and viral genes [21] . This progress bodes well for the future quantification of the MCP using microbial functional genes (Fig. 2).
Large-scale manipulation experiments
The global ocean is experiencing a slowdown in overturning circulation, increasing temperature, decreasing pH, increasing UV radiation, increasing dissolved CO2, decreasingdissolved O2 and changing inorganic and organic nutrient availability [22] , with as-yet unknown consequences for ocean carbon storage. A combination of laboratory microcosm- (10-3 to 1 m3),mesocosm- (1 to 103 m3) and macrocosm- (> 103 m3) scale manipulation experiments are required to test hypotheses related to the individual and combined effects of these multiple drivers on the MCP [23]. Mesocosms deployed at sea and macrocosms on land have been used successfully to investigate the effects of ocean acidification and nutrient additions (via, for example, atmospheric dust deposition) on marine plankton activity [24]. The range of scale is required due to the trade-off between numerous replication and full factorial matrices available at the microcosm scale and improved representation of the complexity and depth distribution of the microbial community, including the potential formation of a pycnocline, at the macrocosm scale [23,25].
Future studies of the MCP should use at-sea mesocosms and on-land macrocosms to bridge the gap between data derived from microcosm bioassay experiments and in-situ measurements. A project to build a facility with 9 X 50 m tall macrocosms to allow experiments to be performed with three replicate controls and three replicates of each of two treatments is currently underway in Qingdao, China [25]. This advanced experimental system will allow the concurrent determination of microbial diversity, the chemical composition of DOC and the rate processes affecting the production and loss of RDOC over relevant space scales of euphotic zone depths and time scales of years, under two environmental treatments in a previously unobtainable statistically robust manner [23]. In addition to advances in process understanding, experiments conducted in meso- and macrocosms can be used as a basis for systematic comparison of methods used elsewhere (components (i) through (iv) above). The design of experiments should be linked directly, when possible, with hypotheses and process-related questions that emerge from field studies, including time-series studies of naturally variable systems (component (vii) above).
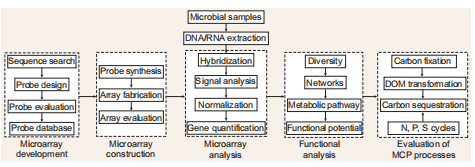
Figure 2. An experimental microarrayprocedureto linkgene analysiswiththequantification of MCP processes.
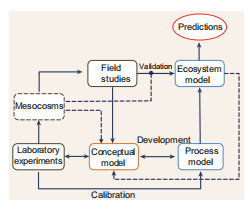
Figure 3. Integration of laboratory, field and numerical studies.
Modelling framework
The dynamics underpinning the MCP are rarely considered in marine ecosystem models and this limits our capacity to predict carbon sequestration in future oceans [26]. While exploratory modelling studies on the MCP have been published recently [26,27], we need to embed these studies in a coherent framework if we want to have reliable MCP simulations. We propose that, in future projects, each aspect of the model development (formulation, calibration, validation) is carried out in co-ordination with empirical studies, in order to exploit the multiple feedbacks between data and simulations to iteratively refine the model and achieve reliable MCP predictions (Fig. 3). Laboratory experiments (i.e. microcosm and mesocosm batch cultures and chemostats) using the techniques described earlier are able to provide a detailed description of RDOC production and microbial consumption in a controlled environment. The understanding generated by these experiments allows the formalization of a conceptual model (i.e. a diagram describing the process being investigated). The conceptual model can be seen as the central hypothesis driving subsequent modelling steps. The conceptual model then needs to be ‘translated' into a mathematical form (process model) and calibrated to reproduce the dynamics observed in the experiments. The development of the process model might highlight knowledge gaps and inform new, more informative, experimental designs. When the process model successfully reproduces the dynamics observed in the experiments, it is ready to be implemented in a full ecosystem model. Field studies (including incubation experiments) provide measurements (standing stock concentrations and rates) to scale the conceptual model up to the ecosystem level and to validate the ecosystem model. Eventually, after the ecosystem model is satisfactorily validated, it can be used for climate predictions. During the validation of the ecosystem model, it is possible to highlight knowledge gaps and inconsistencies which imply that the conceptual model should be revised and new experiments planned. Mesocosms and macrocosms represent a valuable compromise between the laboratory and the real environment, providing additional information to refine the conceptual model, inform the development of the process model and validate the ecosystem model.
Global and decadal scale perspective
In order to predict future changes in the capacity and efficiency of the MCP, global spatial-scale and decadal temporal-scale surveys of robust, high-throughput proxies of RDOC accumulation will be required alongside chemical fingerprints of RDOC transformation pathways, such as D- enantiomers of amino acids, and the more analytically challenging comprehensive chemical characterization of DOC. High-throughput proxies might include the fluorescence intensity of humic-like chromophoric dissolved organic material (CDOM), the production of which correlates with apparent oxygen utilization (i.e. microbial degradation of organic material) in the deep ocean.
Global and decadal monitoring of the MCP would require the development of sensors for components of the DOM pool (e.g. CDOM, fDOM, DOC, RDOC fingerprints) appropriate for deployment on a range of platforms including moorings, gliders, volunteer observing ships (such as ferries and container ships) and biogeochemical ARGO floats (http:// www.argo.ucsd.edu/), as well as remote sensing algorithms for surface distributions. An international co-ordinated programme would be required, including standardized methods, inter-calibration exercises, and scientific training and capacity building. This should be embedded within established time-series stations such as the Hawaii Ocean Timeseries (HOT; http://hahana.soest. hawaii.edu/hot/) and those within the OceanSITES (http://www.oceansites. org/index.html) worldwide network, as well as global shipboard surveys such as those co-ordinated within GO-SHIP (http://www.go-ship.org/ index.html) and GEOT^ACES (http://www.geotraces.org/) to ensure that appropriate biogeochemical, biodiversity and omics data are collected concurrently. The Global Ocean Observing System (GOOS; http://www.goosocean.org/) Biogeochemistry Panel has recognized the need for sustained ocean observations of DOC, designating it as an essential ocean variable (EOV) required for addressing societal drivers such as the role of biogeochemistry in climate and human impacts on ocean biogeochemistry. The future aspiration should be to designate RDOC or a proxy of DOC lability as an EOV.
Progress in MCP quantification will also rely on continued advances in quantification of the microbial processes related to DOC and RDOC production and transformation, including heterotrophic bacterial respiration, viral lysis and chemoautotrophy. Improvements in bioinformatics and data manipulation and visualization tools will enhance the opportunities for connections to bemade between the molecular complexity of RDOC and meta-genomics, -proteomics and -transcriptomics [20].
Given the complexity and diversity of measurements involved (i-iii) and the value of repeated measurement and experimentation for model validation (vii), a logical starting point for a systematic MCP field study would be the addition of MCP appropriate measurements at the locations of already established moored- and ship-based multidisciplinary ocean time series that examine of ocean ecosystems and biogeochemical conditions. Overall, MCP quantification would be most efficiently achieved through a co-ordinated international research initiative linking each of these programme components (i-vii) and including appropriate policies for method standardization and intercalibration, data-management, capacitybuilding, outreach and education, and science-policy-society activities to address the societal driver of understanding the role of marine biogeochemistry in climate (Fig. 4).
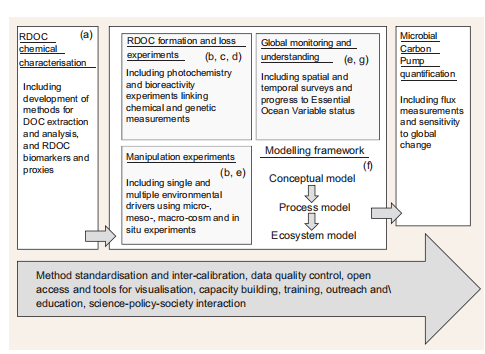
Figure 4. Schematic of a co-ordinated international research initiative linking each of these programme components (i-vii) with data-management, capacity-building and science-policy-society activities.
CONCLUSION
Quantitative understanding of the mechanisms that produce and utilize RDOC is still in its infancy, despite their fundamental role in global climate. Significant progress towards quantification of the MCP requires an internationally co-ordinated multidisciplinary research initiative that interlinks in-situ time-series studies and micro- and macro-scale manipulation experiments within a robust conceptual to numerical modelling framework. The analytical tools and intellectual interaction between chemists and microbiologists are now at a stage where this societally driven question can, and must, become a high priority research topic.
ACKNOWLEDGEMENTS
We sincerely thank the participants of the first Yan-qi Lake conference and the joint International Council for the Exploration of the Sea/North Pacific Marine Science Organisation (ICES/PICES) working group on Climate Change and Biologically-driven Ocean Carbon Sequestration (WGCCBOCS; http://www.ices.dk/community/ groups/Pages/WGCCBOCS.aspx, http://meet ings.pices.int/members/working-groups/wg33).
FUNDING
This work was supported by the United Kingdom Natural Environment Research Council (NERC) (NE/R000956/1 awarded to C.R.), the Lever- hulme Trust (RPG-2017-089 awarded to C.R.), the Korean Long-term Marine Ecological Research (K-LTMER) Program funded by the Korean Ministry of Oceans and Fisheries (awarded to J.-H.- H.) and the United States of America National Science Foundation (1233373 awarded to R.B.). L.P. was funded by the UK NERC National Capability for Marine Modelling, Y.Z. by the National KeyResearch and Development Programs of China (2016YFA0601400)and D.W. was supported by the Canada Excellence Research Chair in Ocean Science and Technology.
Carol Robinson1,*, DouglasWallace2,
Jung-Ho Hyun3, Luca Polimene4, Ronald Benner5, Yao Zhang6, Ruanhong Cai6, Rui Zhang6 and Nianzhi Jiao6
1 University of East Anglia, School of Environmental Sciences, UK
Oceanography Department, Dalhousie University, Canada
3Hanyang University, Department of Marine Science and Convergence Engineering, South Korea
4Plymouth Marine Laboratory, Prospect
Place, UK
5University of South Carolina, Department of
Biological Sciences and School of the Earth, Ocean and Environment, USA
6State Key Laboratory of Marine Environmental Science and Institute of Marine Microbes and Ecospheres, China
* Corresponding author.
E-mail: carol.robinson@uea.ac.uk
REFERENCES
1.Jiao N, Herndl GJ and Hansell DA et al. Nat Rev Microbiol 2010; 8: 593-9.
2.Legendre L, Rivkin RB and Weinbauer MG et al. Prog Oceanogr2015; 134: 432-50.
3.Shen Y and Benner R. SciRep2018; 8: 2542.
4.Jiao N, Robinson C and Azam F et al. Biogeosciences 2014; 11: 5285-306.
5.Hertkorn N, Harir M and Koch BP et al. Biogeosciences 2013; 10:1583-624.
6.Li Y, Harir M and Uhl J. Water Research 2017; 116: 316-23.
7.del Giorgio P and Davies J. Patterns of dissolved organic matter lability and consumption across aquatic ecosystems. In: Findlay SEG and Sins- abaugh RL (eds). Aquatic Ecosystems: Interactivity of Dissolved Organic Matter. San Diego, CA: Academic Press, 2003, 399-424.
8.L0nborg C and Alvarez-Salgado XA. Global Bio- geochem Cycles 2012; 26: GB3018.
9.Riebesell U, Fabry VJ and Hansson L etal. (eds). Guide to Best Practices for Ocean Acidification Research and Data Reporting. Luxembourg: Publications Office ofthe European Union,2010,260.
10.KaiserKandBennerR. LimnolOceanogr2008; 53: 99-112.
11.Benner R and Herndl GJ. Bacterially derived dissolved organic matter in the microbial carbon pump. In: Jiao N, Azam F and Sanders S (eds). Microbial Carbon Pump in the Ocean. Washington, DC: Science/AAAS, 2011,46-8.
12.Dittmar T. Reasons behind the long-term stability of dissolved organic matter. In: Hansell DA and
Carlson CA (eds). BiogeochemistryofMarine Dissolved Organic Matter. Oxford: Academic Press, 2015, 369-88.
13.Arrieta JM, Mayol E and Hansman RL et al. Science 2015; 348: 331-3.
14.Wilson JD and Arndt S. Global Biogeochem Cycles 2017; 31:775-86.
15.Radajewski S, Ineson P and Parekh NR etal. Nature 2000; 403: 646-9.
16.Jiao N and Zheng Q. ApplEnviron Microbiol2011; 77: 7439-44.
17.Tu Q, Yu H and He Z et al. Mol Ecol Resour 2014; 14: 914-28.
18.Wang Y, Zhang R and He Z et al. Front Microbiol 2017; 8: 1153.
19.Moran MA, Kujawinski EB and Stubbins A et al. Proc Natl Acad Sci USA 2016; 113: 3143-51.
20.Reed DC, Algar CK and Huber JA et al. Proc Natl Acad Sci USA 2014; 111: 1879-84.
21.Guidi L, Chaffron S and Bittner L et al. Nature 2016; 532: 465-70.
22.Portner H-O, Karl DM and Boyd PW et al. Ocean systems. In: Field CB, Barros VR and Dokken DJ et al. (eds). Climate Change 2014: Impacts, Adaptation and Vulnerability. Part A: Global and Sectoral Aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, UK: Cambridge University Press and New York, NY: USA, 2014; 411-84.
23.Legendre L, Rivkin RB and Jiao N. ICES J Mar Sci 2018; 75: 30-42.
24.Riebesell U, Bellerby RGJ and Grossart H-P et al. Biogeosciences 2008; 5: 1157-64.
25.Jiao N, Azam F and Legendre L. Marine Ecosystem Experimental Chamber System (MECS)一a powerful tool for scenario studies on climate and environmental changes. 3rd International Symposium, Effects of Climate change on the World's Oceans, March 21-27, 2015, Santos, Brazil, poster W3-G2-P18,2015,273 http://www. pices.int/publications/book_of_abstracts/2015- Brazil-Cimate-Change-Book-of-Abstracts.pdf.
26.Polimene L, Rivkin RB and Luo YW et al. Modelling marine DOC degradation time scales. Natl Sci Rev 2018; 5: 468-74.
27.Polimene L, Sailley S and Clark D etal. J Plankton Res 2017; 39:180-6.
National Science Review
5: 474-480, 2018
doi: 10.1093/nsr/nwy070
Advance access publication 7 July 2018