Unveiling the enigma of refractory carbon in the ocean
Nianzhi Jiao1, Ruanhong Cai1, Qiang Zheng1, Kai Tang1, Jihua Liu2,3, Fanglue Jiao3,Douglas Wallace3, Feng Chen2,4, Chao Li5, Rudolf Amann6, Ronald Benner7 and Farooq Azam8
INTRODUCTION
The ocean holds a tremendous reservoir of refractory dissolved organic carbon (RDOC) that plays an important role in carbon cycling and climate change[1].
However, the origin of the RDOC has been an enigma for half a century. This perspective is to address why the enigma deserves scientific efforts and illustrate a robust scheme一the Microbial Carbon Pump (MCP)一to unveil the enigma from molecular to ecosystem levels. Through generation of intrinsic RDOC (RDOCt) under specific biotic and abiotic environmental conditions, as well as through derivation of diverse organic molecules at extremely low concentrations (RDOCc), the MCP links the seemingly contrary ‘intrinsic recalcitrance hypothesis' and ‘dilution hypothesis' together, and provides a framework for testable hypotheses linking microbial activities with the behavior of organic compounds for future studies regarding carbon sequestration in the ocean.
AHALF-CENTURY ENIGMA
The enigma: how is the huge ocean organic carbon reservoir formed?
The ocean harbors a vast reservoir of RDOC that is equivalent in amount (670 Pg C) to the total inventory of CO2 in the atmosphere [2]. The RDOC can be sequestered in the deep ocean for 4-6 kyr, playing an important role in carbon [2]cycling and climate change [1] . This carbon reservoir was recognized half a century ago and how it is formed has been an intriguing topic for research for decades [3—7]. Given its old age, the RDOC was once attributed to seabed seepage of organics, butlaterstudies on oil decomposition, particularly the consequences of the Deepwater Horizon oil spill, showed that most of the spilled old organic carbon is actually labile for the microbes in the water column [8]. Furthermore, a substantial portion of the RDOC was found recently to have a modern radiocarbon age [9]. While radiocarbon age may not necessarily be a proper proxy of recalcitrance of organic matter, the recalcitrance itself could also be apparent. From this viewpoint, RDOC could be composed of diverse labile compounds at extremely low individual concentrations and thus inaccessible to microbes (i.e. the ‘dilution hypothesis') [3-6], although dilute labile molecules appear to account for a relatively small portion [10,11]. These paradoxes show the complexity of the ocean RDOC and its cycling, and why its origin has remained an enigma for half a century [4,7].
A robust scheme: the MCP
The MCP conceptual framework was proposed to address the mechanisms involved in the formation of RDOC [12]. Three pathways of the MCP are identified: RDOC generated directly from microbial cell materials, RDOC derived from degradation of particulate organic matter and residual RDOC after microbial utilization of the bulk DOC [13]. All RDOC compounds can be classified into two types: RDOCt that is composed of compounds that are intrinsically refractory in a specific environmental context (including its biotic and abiotic aspects) and RDOCc that is composed of a tremendous diversity of labile compounds with extremely low individual concentrations, lying below their uptake thresholds [5]. Products from all three MCP pathways contribute to either RDOCt or RDOCc in all cases (Fig. 1).
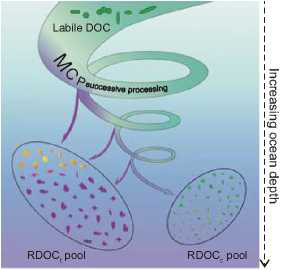
Figure 1. MCP successive processing of organic carbon generates RDOCt as well as RDOCc in the water column. The RDOCt pool is composed of those compounds that are re- fractoryunderagiven environmental context including its biotic and abiotic aspects; RDOCc is composed of diverse labile compounds with extremely low individual concentrations below their uptake thresholds.
TACKLING THE ENIGMA IN THE MCP FRAMEWORK
Molecular characteristics and microbial origin of RDOC
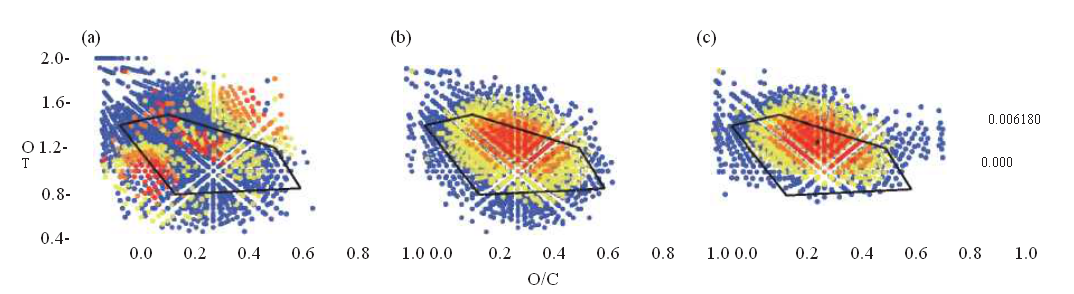
Advances in analytical chemistry, such as ultra-high-resolution Fourier transform-ion cyclotron resonance-mass spectrometry (FT-ICR-MS), high field nuclear magnetic resonance (NMR) spectrometry and excitation emission matrix (EEM) fluorescence have helped to characterize molecular fingerprints and structures of organic compounds in the deep sea. Carboxyl-rich alicyclic molecules (C^AM), characterized by FT-ICR-MS and NMR as refractory components, are found to be widespread in the deep ocean [14]. A large suite of C HO-containing molecules with C^AM-like characteristics (located in the elemental ratio region known as the ‘island of stability', IOS) are reported to have residence times that greatly exceed the oceanic mixing time in the deep Atlantic Ocean [15]. These C^AM-like components of RDOC appear to be oxidized molecules of microbial origin [16,17]. A long-term incubation experiment employing the Aquatron Tower Tank (>100 tons of water) showed robust evidence that phytoplankton debris (Fig. 2a) is effectively transformed to RDOCt during this experiment as indicated by the C^AM pattern (Fig. 2b). These, in turn, resemble the deep-sea sample from the South China Sea (Fig. 2c), indicating that deep-sea RDOC was generated through the MCP.
Observed microbial transformation of organic carbon in the field
Humic-like fluorescent dissolved organic matter (FDOM) is another indicator of the microbial transformation of organic matter, which can be readily recorded in field samples. The turnover time of FDOM is much longer than that of the dark global-ocean circulation [18], indicating that humic-like FDOM is a fraction of the RDOC. FDOM is produced in situ and accumulates in the interior of the ocean. Recent studies have provided new evidence that both cyanobacteria and heterotrophic bacteria contribute to deep-ocean FDOM [19]. These findings not only demonstrate the existence of the MCP, but also suggest rapid microbial modification of the organic carbon structure and its chemical complexity. That is why the dual concepts of RDOCt and RDOCc are necessary [5]. RDOCt varies significantly between surface and deep waters in terms of molecular structures, redox state, degradation state and contributions of diverse compound groups [11,15]. RDOCt in the deep ocean has been modified by microbial enzymatic processes and features relatively higher double-bond equivalent values and degradation state but lower H/C ratios, as well as higher degree of unsaturation plus rings in molecules [20]. In contrast, the majority of the molecular formulas enriched in surface seawater are classified either as highly unsaturated compounds or as unsaturated aliphatic compounds, which make a particularly important contribution at the depth of the chlorophyll maximum, suggesting that these compounds are a major fraction of labile phytoplankton exudates [11]. Only a tiny fraction of the deep-sea organic compounds, on the other hand, are unsaturated aliphatics [11], indicating the existenceof RDOCc in the deep ocean. Compared to the sharp decline in labile DOC compounds with water depth, including unsaturated aliphatic compounds containing nitrogen, the RDOCt such as polycyclic aromatics (PCAs), highly aromatic compounds and highly unsaturated compounds increased with depth [11].
These spatial patterns help to reveal the active transformation of the MCP ongoing within the ocean's water column. The downward transport of young (recently formed) RDOC compounds from the surface ocean to depths by the ocean's ‘mixed-layer pump' [21] as well as the supply of RDOC derived from decomposition of sinking organic particles explains why a considerable fraction of deepocean RDOC (10-30%) has a modern radiocarbon age [9]. On the other hand, there is also ‘recent' formation of RDOC with old radiocarbon signatures ongoing.
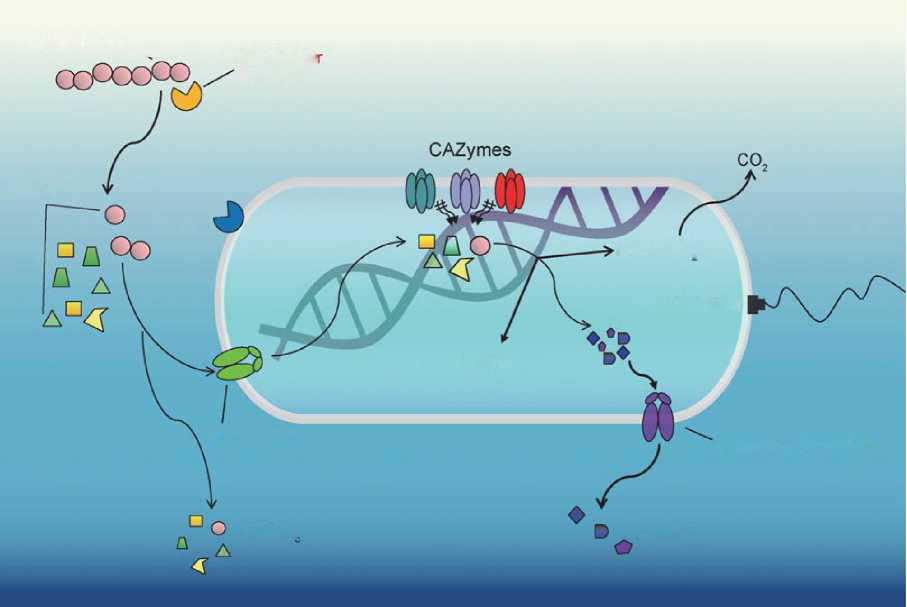
Figure 3. The processes of bacterial hydrolysis, transport, metabolism and secretion of DOC compounds. HMW DOC compounds are degraded by extracellular hydrolyses into LMW DOC molecules and part ofthem are then transported into the cell.There are three fatesfor intracellular metabolism of DOC: energy generation and CO2 release; biosynthesis as cell biomass; and transformation and secretion to the environment, with part of it as RDOCt and RDOCc. CAZymes, the carbohydrate-active enzymes and associated carbohydrate-binding modules involved in the synthesis and degradation of complex carbohydrates.
within the deep sea :e ven if natural hydrocarbon seepage, as well as methane, is labile and capable of rapid remineraliza tion into CO2 [22], subsequent chemosynthetic microbial activity could synthesize the aged CO2 within deep-sea environments such as hydrothermal systems [23], thereby contributing to the RDOC pool via the MCP.
Mechanisms at the cellular, enzymatic and genetic levels
Marine microbiomes are diverse in terms of their community composition and metabolic potentials. In the upper ocean, -20 Gt of carbon is fixed annually by phytoplankton and rapidly metabolized mostly by heterotrophic microbes. Different microbial groups (e.g. Fl^vobacteriia, Roseobacter and Al- teromonas) display a successive order in utilization of the phytoplankton biomass composed primarily of polysaccharides and proteins [24] which consists of hundreds of different compounds [7]. Such successive mineralization processes make use of ectoenzymes, exoenzymes or alternative selfish-uptake mechanisms [24] generating specific RDOCt and RDOCc under corresponding specific environmental contexts [5]. The pathways and rates of microbial transformation determine the fate and the amount of carbon that is converted ultimately to CO2 or RDOC [25]. Currently, the big data approaches of 'omics' (genomics, trans crip tomics, proteomics and metabolomics) open up possibilities to study the transporters and enzymes involved in phytoplankton mineralization and RDOC generation in the natural environment (Fig. 3).
Genomic data analysis can provide information on which microbes utilize what DOC components and thus potentially discriminate RDOCt, while metabolomics information analysis together with advanced FT-ICR-MS and NMR analysis offer an approach for the detection of molecular formulas and chemical structures of RDOCt and RDOCc [5]. Molecular mechanisms for DOC utilization vary among different bacterial groups. For example, ABC transporters responsible for the uptake of low-molecular-weight (LMW) molecules are rich within the Roseobac- ter genomes, while TonB-dependent transporters for high-molecular-weight (HMW) molecules are abundant in the Flavobacteriia genomes [26]. To date, there is a wide variety of small molecules (-14 000) in the biochemical network of microbes, and the HMW molecules derived from macromolecule metabolism (e.g. peptides and peptidoglycan) can also contribute greatly to metabolic diversity. However, there are -600 known transporter reactions, most of which are responsible for transporting a specific substrate or a group of substrates with similar chemical structures. Thus, many metabolites may have no corresponding transporters. Furthermore, some metabolites are produced by known metabolic reactions of an organism and have no reactions consuming it, accounting for -3% of the compounds derived from all pathway reactions of a microbe [27]. The aerobic anoxygenic phototrophic bacterium Roseobacter denitrificans OCh 114 produces oxidized carotenoids spheroiden-2-one and 2, 2'-dioxospirilloxanthin by spheroidene monooxygenase. It becomes obvious that the number of metabolites found in a single species exceeds the number of genes encoding enzymes involved in their uptake and further biotransformation. There are at least 100 000 different compounds in DOM in the deep ocean [28]. The huge gap between the diversity of enzymes and the diversity of DOC compounds likely facilitates the formation and stability of deep-sea RDOCt. On the other hand, the affinity of transporters constants (Ks) varies greatly from millimolar to nanomolar, and are generally higher than the apparent picomolar concentrations of deep-sea compounds [28], consistently with the existence of RDOCc.
FUTURE PERSPECTIVES MCP sustains the global-ocean RDOC pool
Efficient microbial transformations of labile substrates into RDOCt have been demonstrated by both laboratory experiments and field observations [16,29]. A recent study estimates that the annual production of RDOC via the MCP ranges from 0.1 to 0.2 Pg C [30], which is in amount equivalent to 4-8% of the current net annual uptake of atmospheric CO2 by the ocean. At least 25% of the oceanicRDOC is ofbacterial origin [31]. Even with the lowest microbial transformation efficiency (<0.4% of the net marine community production is shunted to RDOC), the MCP can still sustain the ocean's huge global RDOC pool [32]. In addition to sequestration of carbon from the atmosphere, it has been suggested that chemosynthetic crustal microbial communities synthesize DOC from inorganic carbon contained in ridge-flank fluids. This may support an indigenous biosphere that can export substantial fixed carbon to the overlying water columns via the ridge-flank circulation [23]. Furthermore, the isotopic biogeochemical record suggests a huge DOC reservoir in the late Neoproterozoic oceans (>2-3 orders of magnitude more abundant than at present) [1] when metazoans were not evolved but microbes thrived, suggesting that the MCP could have worked very efficiently in ancient times [5]. Recent studies further suggest that the proximal- to-distal marine redox gradient may have favored an intense MCP in shelf areas and the buildup of RDOC in deeper waters of the late Proterozoic oceans [33] . Taken together, the MCP is the principal mechanism generating andsustaining the tremendous oceanic RDOC reservoir.
Future studies
There is an urgent andoverall needto better understand the impacts of global-scale environmental change, including ocean warming and ocean acidification on carbon cycling within the ocean. Improved understanding of the microbial processes responsible for transformations of organic carbon is central to this requirement, among all related issues. In order to advance understanding, in-depth and coordinated studies are particularly required using the following approaches:
Approach 1) Investigation of microbiomes in different, natural environments, including much better coverage of the deep ocean. The Global Ocean Sampling (GOS) expedition only sampled from the surface ocean and the ambitious Tara Oceans expedition collected microbial samples from only three depths (5, 70 and 600 m) [34]. The deeper-ocean microbes are reported to be abundant in metabolic genes related to the degradation of complex organic molecules but there is a lack of sufficient studies. Application of various omics technologies (i.e. metagenomics, metatranscrip- tomics and metaproteomics, and metabolomics) at all levels of the microbial community (i.e. virus, bacteria, archaea, phytoplankton and chemoautotrophs) is needed. In addition to surveys, these methods should also be applied at selected time-series locations in the coastal and open ocean, in order to identify how the metabolic capacity of the ocean's microbiome responds to temporal changes in environmental context.
Approach 2)Linkages between microbial metabolisms and the chemical structure of DOC compounds requires further investigation. In addition to bioassays on changes of DOC composition coupled with changes in bacterial communities, the combination of omics and FT-ICR-MS and NMR technologies offers the potential for new insights into mechanisms responsible for the formation of RDOCt and RDOCc. This area of research is often restricted by technical limitations on DOC extraction and analysis. Coupling of various DOC extraction methods with promising new analytical chemistry approaches is urged to achieve comprehensive understanding on the still largely unknown fingerprints and structures of complex DOC. This is likely to benefit from coordinated experimentation and intercomparison of diverse analytical approaches by different groups working in this area.
Approach 3) A key requirement for making progress in understanding is the establishment and expansion of long-term incubation studies employing large-scale facilities, such as the Aquatron Tower Tank (12 m in height, 3.8 m in diameter) or the MECS (marine environmental chamber system, designed as 50 m in height and 5 m in diameter) under controlled environmental conditions. These facilities offer a unique complement to field studies by allowing the deliberate creation of ocean-relevant physical/ chemical/biological environmental context (e.g. depth profiles) for long-term experimentation. Long-term experiments under controlled yet realistic environmental conditions are required to provide data and insight for testing of theories and hypotheses regarding the effects of global environmental change on ocean carbon cycle. Together with time-series studies, such large-scale experimental facilities also provide opportunities for testing and inter-comparison of diverse and novel analytical and experimental approaches (see Approaches (1) and (2) above). By allowing control of euphotic/aphotic depths, compensation depths, stratification, nutrient gradients, oxygen levels and redox gradient, pH, etc., large-scale experimental facilities can enable focused and repeatable studies of the microbial processing of organic carbon and its role for the consequent transformation and sequestration of carbon within the ocean.
ACKNOWLEDGEMENTS
We thank the participants of the first Yanqi Lake conference and the PICES-ICES WG on Biological Driven Carbon Pumps; Dr. Quan Shi and Chen He from China University of Petroleum-Beijing forthe assistance in FT-ICR-MS data analysis; the Aquatron team led by John Batt at Dalhousie University. This work was supported by the National Key Research Programs (2016YFA0601100), National Program on Global Change and Air-Sea Interaction (GASI-03-01-02-05), the National Natural Science Foundation of China project (91751207 and 91428308), the Chinese Academy of Sciences consulting project(2016ZWH008A-008)and the NSFC-CAS joint project on discipline strategy (L1624030).
Nianzhi Jiao1,*, Ruanhong Cai1, Qiang Zheng1, Kai Tang1, Jihua Liu2,3, Fanglue Jiao3, DouglasWallace3, Feng Chen2,4, Chao Li5, RudolfAmann6, Ronald Benner7 and Farooq Azam8 1 State Key Laboratory of Marine Environmental Science and Institute of Marine Microbes and Ecospheres, Xiamen University, China 2Institute of Marine Science and Technology, Shandong University, China 3Oceanography Department, Dalhousie University,Canada 4Environmental Research Center, University of Maryland at Baltimore, USA 5State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences, China 6Max Planck Institute for Marine Microbiology, Germany 7Department of Biological Sciences and Marine Science Program, University of South Carolina, USA 8Scripps Institution of Oceanography, University of California, USA
* Corresponding author.
E-mail: jiao@xmu.edu.cn
REFERENCES
1.Rothman DH, Hayes JM and Summons RE. Proc NatlAcad Sci USA 2003; 100: 8124-9.
2.Hansell DA, Carlson CA and Repeta DJ et al. Oceanog 2009; 22: 202-11.
3.Jannasch HW. Limnol Oceanogr 1967; 12: 26471.
4.Barber RT. Nature 1968; 220: 274-5.
5.Jiao N, Robinson C and Azam F et al. Biogeosciences 2014; 11: 5285-306.
6.Arrieta JM, Mayol E and Hansman RL et al. Science 2015; 348: 331-3.
7.Moran MA, Kujawinski EB and Stubbins A et al. Proc Natl Acad Sci USA 2016; 113: 3143-51.
8.Chanton JP, Cherrier Jand Wilson RM etal. Environ Res Lett 2012; 7: 045303.
9.Follett CL, Repeta DJ and Rothman DH et al. Proc Natl Acad Sci USA 2014; 111: 16706-11.
10.Jiao NZ, Legendre L and Robinson C et al. Science 2015; 350: 1483.
11.Medeiros PM, Seidel M and PowersLC etal. Geo- physResLett2015; 42: 863-70.
12.Jiao N, Herndl GJ and Hansell DA et al. Nat Rev Micro 2010; 8: 593-9.
13.Jiao N and Azam F. Microbial carbon pump and its significance for carbon sequestration in the ocean. In: Jiao N, Azam F and Sanders S (eds). Microbial Carbon Pump in the Ocean. Washington, DC: Science/AAAS, 2011, 43-5.
14.Hertkorn N, Harir M and Koch BP et al. Biogeosciences 2013; 10: 1583-624.
15.Lechtenfeld OJ, Kattner G and Flerus R et al. Geochim Cosmochimica Acta 2014; 126: 321-37.
16.Lechtenfeld OJ, Hertkorn N and Shen Y etal. Nat Commun 2015; 6: 6711.
17.Koch BP, Kattner G and Witt M et al. Biogeosciences 2014; 11: 4173-90.
18.Catala TS, Reche I and Fuenteslema A etal. Nat Commun 2015; 6: 5986.
19.Zhao Z, Gonsior M and Luek J et al. Nat Comms 2017; 8: 15284.
20.Koch BP and Dittmar T. Rapid Commun Mass Spectrom 2006; 20: 926-32.
21.Dall'Olmo G, Dingle J and Polimene L etal. Nature Geosci2016; 9: 820-3.
22.Pohlman JW, Bauer JE and Waite WF et al. Nature Geosci 2011; 4: 37-41.
23.McCarthy MD, Beaupre SR and Walker BD et al. Nature Geosci 2011; 4: 32-6.
24.Teeling H, Fuchs BM and Becher D et al. Science 2012; 336: 608-11.
25.Ann MM and Hodson RE. Limnol Oceanogr 1994; 39: 762-71.
26.Tang K, Jiao N and Liu K et al. PLoS ONE 2012; 7: e41204.
27.Mackie A, Keseler IM and Nolan L et al. PLoS ONE 2013; 8: e75210.
28.Zark M, Christoffers J and Dittmar T. Mar Chem 2017; 191:9-15.
29.Ogawa H, Amagai Y and Koike I et al. Science 2001; 292: 917-20.
30.Legendre L, Rivkin RB and Weinbauer MG et al. Prog Oceanogr 2015; 134: 432-50.
31.Kaiser K and Benner R. Limnol Oceanogr 2008; 53: 99-112.
32.Osterholz H, Niggemann J and Giebel HA et al. Nat Commun 2015; 6: 7422.
33.Li C, Hardisty DS and Luo G et al. Geobiology 2017; 15: 211-24.
34.Sunagawa S, Coelho LPand Chaffron S etal. Science 2015; 348: 1261359.
National Science Review
5: 459-463, 2018
doi: 10.1093/nsr/nwy020
Advance access publication 5 February 2018

